For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TSRGISRSTLLALWGVFLALPVAATMLYSVATVWRGRALPDGFTVHWWVQTLSEPRVVAALLRSTWLATL TVLIVAAVVLPALYWGYIRNPRIRTTMQVCALLPFALPFVVLAYGIKRLAGATEVTQPWEASPLLVVLGH VALAFPFFLWPVDGAMAAAGVRNLSEAAETSGASPLSTLFRVVIPNIRTGILTGAILTFATSFGEYSIAR VITGSSFETLPVWQVAALQDNRGNPNGVAVMAMFTFVLMFVVSVLIARTGRGEPVRLLPGINTTQKR*
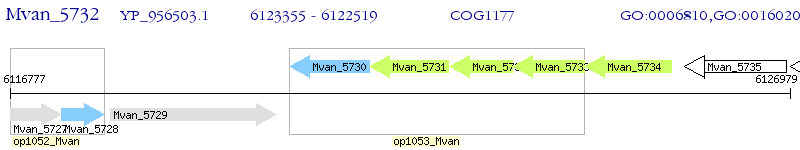
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_5732 | - | - | 100% (278) | binding-protein-dependent transport systems inner membrane component |
| M. vanbaalenii PYR-1 | Mvan_5740 | - | 4e-26 | 29.88% (251) | binding-protein-dependent transport systems inner membrane |
| M. vanbaalenii PYR-1 | Mvan_3390 | - | 4e-11 | 25.73% (241) | binding-protein-dependent transport systems inner membrane |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1889 | modB | 5e-11 | 28.03% (264) | molbdenum-transport integral membrane protein ABC MODB |
| M. gilvum PYR-GCK | Mflv_1082 | - | 5e-26 | 29.48% (251) | binding-protein-dependent transport systems inner membrane |
| M. tuberculosis H37Rv | Rv1858 | modB | 6e-11 | 28.03% (264) | molbdenum-transport integral membrane protein ABC |
| M. leprae Br4923 | MLBr_01088 | - | 0.0001 | 22.52% (222) | putative ABC-transport protein, inner membrane component |
| M. abscessus ATCC 19977 | MAB_1654 | - | 3e-12 | 26.15% (260) | sulfate ABC transporter, permease protein CysW |
| M. marinum M | MMAR_2732 | modB | 5e-10 | 28.19% (227) | molybdenum-transport integral membrane protein ABC |
| M. avium 104 | MAV_2861 | modB | 1e-11 | 30.34% (234) | molybdate ABC transporter, permease protein |
| M. smegmatis MC2 155 | MSMEG_6670 | - | 1e-132 | 83.76% (271) | ABC transporter, permease protein, putative |
| M. thermoresistible (build 8) | TH_3819 | - | 2e-12 | 27.27% (264) | PUTATIVE NifC-like ABC-type porter |
| M. ulcerans Agy99 | MUL_3021 | modB | 4e-11 | 29.07% (227) | molybdenum-transport integral membrane protein ABC |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1889|M.bovis_AF2122/97 ----------------------MHPPTDLPRWVYLPAIAGIVFVAMPLVA
Rv1858|M.tuberculosis_H37Rv ----------------------MHPPTDLPRWVYLPAIAGIVFVAMPLVA
MAV_2861|M.avium_104 --------------------------------MYLPAAAGTMFVVLPLLA
MMAR_2732|M.marinum_M ----------------------MHGPTELPRWVYLPATAGMGLVILPLVA
MUL_3021|M.ulcerans_Agy99 ----------------------MHGPPELPRWVYLPATAGMGLVILPLVA
TH_3819|M.thermoresistible__bu ----------------------MNAST-LPRWIYLPAAVGATLLALPVFA
MAB_1654|M.abscessus_ATCC_1997 ---------------------MTLSPAVKYALRILALLYVAVLLIVPLGL
Mvan_5732|M.vanbaalenii_PYR-1 -----------------MTSRGISRSTLLALWGVFLALPVAATMLYSVAT
MSMEG_6670|M.smegmatis_MC2_155 -----------------MTR--MSRWALLGLWSVFLVLPVLATLLYSLAT
Mflv_1082|M.gilvum_PYR-GCK ---------------------MADVMRLVRGCLWVMFGLFFLFPLYAMAD
MLBr_01088|M.leprae_Br4923 MGIEKVKVTAGVRVGARRATIWVIIDTFVVGYALLPVLWILSLSLKPAST
. .
Mb1889|M.bovis_AF2122/97 IAIRVDWP-----RFWALITT-PSSQTALLLSVKTAAASTVLCVLLGVPM
Rv1858|M.tuberculosis_H37Rv IAIRVDWP-----RFWALITT-PSSQTALLLSVKTAAASTVLCVLLGVPM
MAV_2861|M.avium_104 IAVKVDWP-----HFWSLITS-PSSRTALLLSLRTAAASTALCVALGVPM
MMAR_2732|M.marinum_M MAAKVDWP-----NFWSLITS-TSSRTALVLSLKTASASTAICVLLGVPM
MUL_3021|M.ulcerans_Agy99 MAAKVDWP-----NFWSLITS-TSSRTALVLSLKTASASTAICVLLGVPT
TH_3819|M.thermoresistible__bu LAGRVDWP-----RFWSLICS-ESSMTALALSLRTATASTVLCLVLGVPM
MAB_1654|M.abscessus_ATCC_1997 ILWRAFAPG--LVQFWEYIST-PAAISAFNLSLLVVAIVVPLNVIFGVPT
Mvan_5732|M.vanbaalenii_PYR-1 VWRGRALPDGFTVHWWVQTLSEPRVVAALLRSTWLATLTVLIVAAVVLPA
MSMEG_6670|M.smegmatis_MC2_155 VWRGRAFPDGYTLAWWVQAFSEPRVVSALMRSVWLAVLTVVVVAAVVLPA
Mflv_1082|M.gilvum_PYR-GCK FSTRNLISDGRTWQAWANLVTDDALYRSIVVSLLLAVLTVVAMLVLLVPT
MLBr_01088|M.leprae_Br4923 VKDGKLIPSLVTFDNYRGIFRSDLFSSALINSIGIGLTTTVIAVMFGAMA
. . : :: * . .
Mb1889|M.bovis_AF2122/97 ALVLARSRGRLVRSLRPLILLPLVLPPVVGGIALLYAFGRLGLIGRYLEA
Rv1858|M.tuberculosis_H37Rv ALVLARSRGRLVRSLRPLILLPLVLPPVVGGIALLYAFGRLGLIGRYLEA
MAV_2861|M.avium_104 ALVLARGGTRLVRSLRPLILLPLVLPPVVGGIALLYAFGRLGLLGHYLEA
MMAR_2732|M.marinum_M ALVLARSRARLMRLMRPLILLPLVLPPVVGGIALLYAFGRLGLIGRYLEA
MUL_3021|M.ulcerans_Agy99 ALVLARSRARLMRLMRPLILLPLVLPPVVGGIALLYAFGRLGLIGRYLEA
TH_3819|M.thermoresistible__bu ALALARGGSPILRWIRPLVLLPLVLPPVVGGIALLYAFGRMGLIGGYLEA
MAB_1654|M.abscessus_ATCC_1997 ALVLARNKFRGKGVLQAIIDLPFAVSPVIVGVALIVLWGSAGLLGFVEND
Mvan_5732|M.vanbaalenii_PYR-1 LYWGYIRNPRIRTTMQVCALLPFALPFVVLAYGIKRLAGATEVTQPWEAS
MSMEG_6670|M.smegmatis_MC2_155 LYWGHIRNGRIRTVMQLCALLPFALPFVVLAYGIKRLAGASELTQPWESS
Mflv_1082|M.gilvum_PYR-GCK MIWVRLRAPWAKGLVEFLCLLPLTIPALVIVVGLRNVY--LWVVYFLGES
MLBr_01088|M.leprae_Br4923 AYAIARLAFPGKRLLVGTTLLVTMFPAISLVTPLFNIERRVGLFDTW---
: * .. : : :
Mb1889|M.bovis_AF2122/97 AGISIAFSTAAVVLAQTFVSLPYLVISLEGAARTAGA-DYEVVAATLGAR
Rv1858|M.tuberculosis_H37Rv AGISIAFSTAAVVLAQTFVSLPYLVISLEGAARTAGA-DYEVVAATLGAR
MAV_2861|M.avium_104 AGISVAFSTTAVVLAQTFVSLPFLVISLEGAARTAGA-DFEVVAATLGAR
MMAR_2732|M.marinum_M GGITIAFSTTAVVLAQTFVSLPFLVVSLEGAARTAGA-DYEVVAATLGAR
MUL_3021|M.ulcerans_Agy99 GGITIAFSTTAVVLAQTFVSLPFLVVPLEGAARTAGA-DYEVVAATLGAR
TH_3819|M.thermoresistible__bu VGIQIAFSTTAVVLAQTFVSLPFLVLALEGAARTAGT-GYEVVAATLGAT
MAB_1654|M.abscessus_ATCC_1997 WKFKIIFGLPGIVLASIFVTLPFVIREVEPVLHEIGD-DQEQAAATLGAN
Mvan_5732|M.vanbaalenii_PYR-1 P--------LLVVLGHVALAFPFFLWPVDGAMAAAGVRNLSEAAETSGAS
MSMEG_6670|M.smegmatis_MC2_155 P--------VLVVLGHVALSFPFFLWPVDGAMSAAGVRQLSEAAETSGAT
Mflv_1082|M.gilvum_PYR-GCK A--------LTLTFVYVIVVLPFAYRAIDAALSAIDLQTLSEAARSLGAG
MLBr_01088|M.leprae_Br4923 ---------AGLILPYITFALPLAIYTLSAFFAEIPW-DLEKAAKMDGAT
: : . :* :. . .* **
Mb1889|M.bovis_AF2122/97 PGTVWWRVTLPLLLPGVVSGSVLAFARSLGEFGATLTFAGSRQGVTRTLP
Rv1858|M.tuberculosis_H37Rv PGTVWWRVTLPLLLPGVVSGSVLAFARSLGEFGATLTFAGSRQGVTRTLP
MAV_2861|M.avium_104 PTTVWWRVTLPLLLPGVVSGAVLAFARSLGEFGATLTFAGSRQGVTRTLP
MMAR_2732|M.marinum_M PSAVWWKVTLPLLMPGLVSGAVLAFARALGEFGATLTFAGSRQGVTRTLP
MUL_3021|M.ulcerans_Agy99 PSAVWWKVTLPLLMPGLVSGAVLAFARSLGEFGATLTFAGSRQGVTRTLP
TH_3819|M.thermoresistible__bu PTTVWWRVTLPLLAPGLVSGAVLAFARALGEFGATLTFAGSRQGVTRTLP
MAB_1654|M.abscessus_ATCC_1997 PWQTFWRITLPSIRWGLTYGIVLTVARTLGEYGAVIMVSSGLPGKSQTLT
Mvan_5732|M.vanbaalenii_PYR-1 PLSTLFRVVIPNIRTGILTGAILTFATSFGEYSIARVITGSSFETLPVWQ
MSMEG_6670|M.smegmatis_MC2_155 PLATLFRVVIPNIRTGILTGAILTFATSFGEYSIARVITGSSFETLPVWQ
Mflv_1082|M.gilvum_PYR-GCK WFTTITRVVVPNIWSGILSAAFISIAVVLGEYTIASLSG---YETLQVQI
MLBr_01088|M.leprae_Br4923 SGQAFRKVIVPLAAPGLVTAAILVFIFAWNDLLLALSLTSTKAAITAPVA
. :: :* *: . .: . .: .
Mb1889|M.bovis_AF2122/97 LEIYL-QRVT-DPDAAVALSLLLVVVAALVVLGVG-------ARTPIGTD
Rv1858|M.tuberculosis_H37Rv LEIYL-QRVT-DPDAAVALSLLLVVVAALVVLGVG-------ARTPIGTD
MAV_2861|M.avium_104 LEIYL-QRVT-DADAAVALSILLVAVAAVVVLGLG-------ARRLTGTD
MMAR_2732|M.marinum_M LEIYL-QRVN-DPDAAIALSLLLVAVAALVVLSLG-------ARRLTGGQ
MUL_3021|M.ulcerans_Agy99 LEIYL-QRVN-DPDAAIALSLLLVAVAALVVLSLG-------ARRLTGGQ
TH_3819|M.thermoresistible__bu LEIYL-QREA-DPEAAVALSMVLIAVAAVVVIGLG-------ARRLRAGG
MAB_1654|M.abscessus_ATCC_1997 LLVSD-RHLRGDEYGAYALSVVLMAVALIVLVVQV-------VLDRNRAK
Mvan_5732|M.vanbaalenii_PYR-1 VAALQ-DNRG-NPNGVAVMAMFTFVLMFVVSVLIARTGRGEPVRLLPGIN
MSMEG_6670|M.smegmatis_MC2_155 VAALQ-DTRG-NPNGVAVMAIFTFLLMFIVSVLIARTGKGQPLRLLPGIG
Mflv_1082|M.gilvum_PYR-GCK VLIGK-SDGPTSVAASLATLLFGFALLLILSLVTR-------GRRNVGVT
MLBr_01088|M.leprae_Br4923 ITNFTGSSQFEEPTGSIAAGAIVITAPIIVFVWIFQR------RIVAGLT
: . . . . . :: :
Mb1889|M.bovis_AF2122/97 TR----
Rv1858|M.tuberculosis_H37Rv TR----
MAV_2861|M.avium_104 AR----
MMAR_2732|M.marinum_M AG----
MUL_3021|M.ulcerans_Agy99 AG----
TH_3819|M.thermoresistible__bu WS----
MAB_1654|M.abscessus_ATCC_1997 TSK---
Mvan_5732|M.vanbaalenii_PYR-1 TTQKR-
MSMEG_6670|M.smegmatis_MC2_155 NTR---
Mflv_1082|M.gilvum_PYR-GCK T-----
MLBr_01088|M.leprae_Br4923 SGAVKG