For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VADVMRLVRGCLWVMFGLFFLFPLYAMADFSTRNLISDGRTWQAWANLVTDDALYRSIVVSLLLAVLTVV AMLVLLVPTMIWVRLRAPWAKGLVEFLCLLPLTIPALVIVVGLRNVYLWVVYFLGESALTLTFVYVIVVL PFAYRAIDAALSAIDLQTLSEAARSLGAGWFTTITRVVVPNIWSGILSAAFISIAVVLGEYTIASLSGYE TLQVQIVLIGKSDGPTSVAASLATLLFGFALLLILSLVTRGRRNVGVTT
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
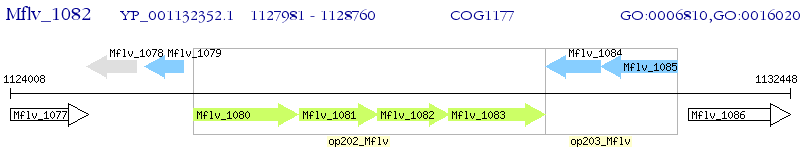
| M. gilvum PYR-GCK | Mflv_1082 | - | - | 100% (259) | binding-protein-dependent transport systems inner membrane component |
| M. gilvum PYR-GCK | Mflv_2679 | - | 2e-12 | 26.05% (215) | sulfate ABC transporter, inner membrane subunit CysT |
| M. gilvum PYR-GCK | Mflv_3135 | - | 5e-10 | 26.12% (245) | binding-protein-dependent transport systems inner membrane |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2421c | cysT | 2e-13 | 28.51% (221) | sulfate-transport integral membrane protein ABC transporter |
| M. tuberculosis H37Rv | Rv2399c | cysT | 2e-13 | 28.51% (221) | sulfate-transport integral membrane protein ABC transporter |
| M. leprae Br4923 | MLBr_02093 | pstA1 | 5e-08 | 28.66% (164) | membrane-bound component of phosphate transport |
| M. abscessus ATCC 19977 | MAB_1504 | - | 2e-14 | 29.70% (202) | ABC transporter permease |
| M. marinum M | MMAR_3717 | cysT | 9e-12 | 26.38% (254) | sulfate-transport integral membrane protein ABC transporter |
| M. avium 104 | MAV_1783 | cysT | 2e-14 | 28.80% (250) | sulfate ABC transporter, permease protein CysT |
| M. smegmatis MC2 155 | MSMEG_6522 | - | 1e-120 | 87.50% (248) | ABC transporter, inner membrane subunit |
| M. thermoresistible (build 8) | TH_1446 | cysT | 6e-15 | 28.29% (251) | PROBABLE SULFATE-TRANSPORT INTEGRAL MEMBRANE PROTEIN ABC |
| M. ulcerans Agy99 | MUL_3660 | cysT | 2e-11 | 26.95% (256) | sulfate-transport integral membrane protein ABC transporter |
| M. vanbaalenii PYR-1 | Mvan_5740 | - | 1e-129 | 92.83% (251) | binding-protein-dependent transport systems inner membrane |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1082|M.gilvum_PYR-GCK --------------------------------MADVMRLVRGCLWVMFGL
Mvan_5740|M.vanbaalenii_PYR-1 ---------------------------------------MRGALWVMFGL
MSMEG_6522|M.smegmatis_MC2_155 -------------------MSTSTTTGTSTGSNRALRRAVRATLWVLFGL
Mb2421c|M.bovis_AF2122/97 -------------MTESLVGER----RAPQFRARL--SGPAGPPSVRVGM
Rv2399c|M.tuberculosis_H37Rv -------------MTESLVGER----RAPQFRARL--SGPAGPPSVRVGM
MAV_1783|M.avium_104 ----------MTAITPDPLAIRPELGGDPGHQPVP--GRGRGTTALRVGV
MMAR_3717|M.marinum_M MTVTAHSGQSDPPAVAPDPGQRAD--GGPQWISGLGRSGRRGATALQVGV
MUL_3660|M.ulcerans_Agy99 MTVTAHSGQSDPPAVAPDLGQRAD--GGPQWISGPGRSGRRGATALQVGV
TH_1446|M.thermoresistible__bu ---------------VSAVLPRSE--TTPAAPPGW---GRGGATSLRVGI
MAB_1504|M.abscessus_ATCC_1997 -----------------------------------MLLWNGRSRLILWSV
MLBr_02093|M.leprae_Br4923 -------------MNLDTLDRPVKAVVLRPISVRRRIKNNVATMFFFTSF
. ..
Mflv_1082|M.gilvum_PYR-GCK FFLFP-LYAMADFSTRNLISDGRTWQ-----AWANLVTDDALYRSIVVSL
Mvan_5740|M.vanbaalenii_PYR-1 FFLFP-LYAMADFSTRNLIAGGRTWR-----AWANLFADQALYQAIVVSL
MSMEG_6522|M.smegmatis_MC2_155 FFLFP-LYAMADFSTRNLISGGRTLA-----AWGNLVADEALYRAIVISL
Mb2421c|M.bovis_AF2122/97 AVVWLSVIVLLPLAAIVWQAAGGGWR-----AFWLAVSSHAAMESFRVTL
Rv2399c|M.tuberculosis_H37Rv AVVWLSVIVLLPLAAIVWQAAGGGWR-----AFWLAVSSHAAMESFRVTL
MAV_1783|M.avium_104 ATVWLSVIVLLPLAAIAWQSAGGGWH-----VFWLAVTSNAALESFRVTL
MMAR_3717|M.marinum_M SMVWLSVIVLLPLAAIAWQAADGGWH-----AFWLSVSSHAALESLRVTL
MUL_3660|M.ulcerans_Agy99 SMVWLSVIVLLPLAAIAWQAADGGWH-----AFWLSVSSHAALESLRVTL
TH_1446|M.thermoresistible__bu ATVWLSIIVLLPLAAILWQSTNGGWD-----AFRDAVTANAALTSLRVTL
MAB_1504|M.abscessus_ATCC_1997 FAVVITVVFIAPIATVVLAGFSGAWTGPLPTQLSFMRYEKALGGDDLASM
MLBr_02093|M.leprae_Br4923 LIALVPLVWLLSVVVARGFYAVTRFDWWT-HSLRGVMPEEFAGGVYHALY
: : . . .
Mflv_1082|M.gilvum_PYR-GCK LLAVLTVVAMLVLLVPTMIWVRLRAPWA----KGLVEFLCLLPLTIPALV
Mvan_5740|M.vanbaalenii_PYR-1 LLAVFTVAAMLVLLVPTMIWVRLRAPWA----KGLVEFMCLLPLTIPALV
MSMEG_6522|M.smegmatis_MC2_155 LLAVFTVVAMLVLLVPTMIWVRLRARWA----RRIVEFLCLLPLTIPALV
Mb2421c|M.bovis_AF2122/97 TISTA-VTVINLVFGLLIAWVLVRDDFAG---KRIVDAIIDLPFALPTIV
Rv2399c|M.tuberculosis_H37Rv TISTA-VTVINLVFGLLIAWVLVRDDFAG---KRIVDAIIDLPFALPTIV
MAV_1783|M.avium_104 TISAG-VTVLNLVFGLVIAWVLVRDEFPG---KRLVDAIIDLPFALPTIV
MMAR_3717|M.marinum_M TVSAG-VTLLNTVFGLLIAWVLIRDEFVG---KRLVDAVIDLPFALPTIV
MUL_3660|M.ulcerans_Agy99 TVSAG-VTLLNAVFGLLIAWGLIRDEFVG---KRLVDAVIDLPFALPTIV
TH_1446|M.thermoresistible__bu TIAAG-VTVVNTVFGLMVAWVLTRDEFIG---KRLVNSVIDLPFALPTIV
MAB_1504|M.abscessus_ATCC_1997 AVSLQTAVMSSGVALLLGTWAALSVREVPSWLRRVIDAVFHLPIAIPSVA
MLBr_02093|M.leprae_Br4923 GSLVQAVVAAIMAVPLGSMTAVYLVEYGSGPFVRVTTFMVDVLAGVPSIV
.. : : : :*::.
Mflv_1082|M.gilvum_PYR-GCK IVVGLRNVY---LWVVYFLGESALTLTFVYVIVVLPFAYRAIDAALSAID
Mvan_5740|M.vanbaalenii_PYR-1 IVVGLRNVY---LWVTYFLGESALTLTFVYVVIVLPFAYRAIDAALSAID
MSMEG_6522|M.smegmatis_MC2_155 IVVGLRNVY---LWVTYFLGESALTLTFVYVVLVLPFAYRAIDSALAAID
Mb2421c|M.bovis_AF2122/97 ASLVMLALYGNNSPVGLHFQHTATGVGVALAFVTLPFVVRAVQPVLLEID
Rv2399c|M.tuberculosis_H37Rv ASLVMLALYGNNSPVGLHFQHTATGVGVALAFVTLPFVVRAVQPVLLEID
MAV_1783|M.avium_104 ASLVMLALYGNNSPVGLHLQHTAAGVAVALAFVTLPFVVRAVQPVLLELD
MMAR_3717|M.marinum_M ASLVMLALYGHNSPLGLHLQHTVWGVGVALAFVTLPFVVRAVQPVLLELD
MUL_3660|M.ulcerans_Agy99 ASLVMLALYGHNSPLGLHLQHTVWGVGVALAFVTLPFVVRAVQPVLLELD
TH_1446|M.thermoresistible__bu ASLVMLALYGPASPVDIHLQHTRWGVGVALLFVTLPFVVRSVQPVLLELD
MAB_1504|M.abscessus_ATCC_1997 IGLGLLTAFN---EPPLLLGGTKWIVIVAHTALVLAFAFSSVSAALERLD
MLBr_02093|M.leprae_Br4923 AALFIFCLW----IATLGFQQSAFAVSLALVLLMLPVVVRSTEEILRLVP
: : : : : : .. : *... : . * :
Mflv_1082|M.gilvum_PYR-GCK LQTLSEAARSLGAGWFTTITRVVVPNIWSGILSAAFISIAVVLGEYTIAS
Mvan_5740|M.vanbaalenii_PYR-1 LQTLSEAARSLGAGWLTTITRVVVPNIWSGILSAAFISIAVVLGEFTIAS
MSMEG_6522|M.smegmatis_MC2_155 LQTLSEAARSLGAGWFTTITRVIVPNIWSGILSAAFISIAVVLGEYTIAS
Mb2421c|M.bovis_AF2122/97 RET-EEAAASLGANGAKIFTSVVLPSLTPALLSGAGLAFSRAIGEFGSVV
Rv2399c|M.tuberculosis_H37Rv RET-EEAAASLGANGAKIFTSVVLPSLTPALLSGAGLAFSRAIGEFGSVV
MAV_1783|M.avium_104 RET-EEAAASLGASGPKIFTSVVLPALLPALLSGAGLAFSRAIGEFGSVV
MMAR_3717|M.marinum_M RET-EEAAASLGATGWKVFTSVIFPSLMPALLSGAGLAFSRAIGEFGSVV
MUL_3660|M.ulcerans_Agy99 RET-EEAAASLGATGWKVFTSVIFPSLMPALLSGAGLAFSRAIGEFGSVV
TH_1446|M.thermoresistible__bu RET-EEAAASLGANNLTIFTRVVLPALLPALLSGAGLAFSRAIGEFGSVV
MAB_1504|M.abscessus_ATCC_1997 PAY-RQVAESLGAGSVRVVRTVTLPLLMPSLGAAAGLAIALSMGELGATV
MLBr_02093|M.leprae_Br4923 DEL-REACYALGIPKWKTIVRIVFPIATPGIISGVLLSIARVIGETAPVL
:.. :** . : .* ..: :.. :::: :** .
Mflv_1082|M.gilvum_PYR-GCK LSG---------------YETLQVQIVLIGKSDGPTSVAASLATLLFGFA
Mvan_5740|M.vanbaalenii_PYR-1 LSG---------------YETLQVQIVLIGKSDGPTSVAASLATLLFGFV
MSMEG_6522|M.smegmatis_MC2_155 LSG---------------FETLQVQIVLIGKSDGPTSVAASLAVLLLGFV
Mb2421c|M.bovis_AF2122/97 LIGGAV--------PGKTEVSSQWIRTLIENDDRTGAAAISVVLLSISFI
Rv2399c|M.tuberculosis_H37Rv LIGGAV--------PGKTEVSSQWIRTLIENDDRTGAAAISVVLLSISFI
MAV_1783|M.avium_104 LIGGAV--------PGKTEVSSQWIRTLIENDDRTGAAAISIVLLAISFV
MMAR_3717|M.marinum_M LIGGAV--------PGETEVSSQWIRTLIENDDRTGAAAISIVLLSISFA
MUL_3660|M.ulcerans_Agy99 LIGGAV--------PGETEVSSQWIRTLIENDDRTGAAAISIVLLSISFA
TH_1446|M.thermoresistible__bu LIGGAV--------PGETEVASQWIRTLIENDDRTGAAAVSIVLLVISFL
MAB_1504|M.abscessus_ATCC_1997 MVYP--------------SSWRTLPVTIFGLSDRGQVFSAAASTTLLLMV
MLBr_02093|M.leprae_Br4923 VLVGYSRSINFDIFDGNMASLPLLIYTELTNPEYAGFLRVWGAALTLIIL
: . : : : :
Mflv_1082|M.gilvum_PYR-GCK LLLILSLVTRGRR-NVGVTT---
Mvan_5740|M.vanbaalenii_PYR-1 LLLILSLVTRGRR-NVGVTV---
MSMEG_6522|M.smegmatis_MC2_155 LLLALAVLTRGRRRTLGDQG---
Mb2421c|M.bovis_AF2122/97 VLLILRVVGARAAKREEMAA---
Rv2399c|M.tuberculosis_H37Rv VLLILRVVGARAAKREEMAA---
MAV_1783|M.avium_104 VLLILRVVGGRAAKREELAA---
MMAR_3717|M.marinum_M VLLVLRVLGARAAKREELAP---
MUL_3660|M.ulcerans_Agy99 VLLVLRVLGARAAKREELAP---
TH_1446|M.thermoresistible__bu VLLVLRVIGSRAAKREEQTA---
MAB_1504|M.abscessus_ATCC_1997 TLLALLVVGRLRGKVAFR-----
MLBr_02093|M.leprae_Br4923 VAGINLFGAAFRFLATRRCSGCR
.