For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MHPPTDLPRWVYLPAIAGIVFVAMPLVAIAIRVDWPRFWALITTPSSQTALLLSVKTAAASTVLCVLLGV PMALVLARSRGRLVRSLRPLILLPLVLPPVVGGIALLYAFGRLGLIGRYLEAAGISIAFSTAAVVLAQTF VSLPYLVISLEGAARTAGADYEVVAATLGARPGTVWWRVTLPLLLPGVVSGSVLAFARSLGEFGATLTFA GSRQGVTRTLPLEIYLQRVTDPDAAVALSLLLVVVAALVVLGVGARTPIGTDTR
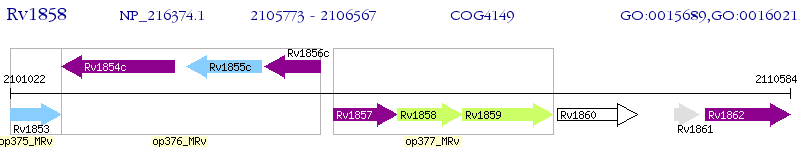
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv1858 | modB | - | 100% (264) | PROBABLE MOLBDENUM-TRANSPORT INTEGRAL MEMBRANE PROTEIN ABC TRANSPORTER MODB |
| M. tuberculosis H37Rv | Rv2398c | cysW | 3e-32 | 35.69% (255) | sulfate-transport integral membrane protein ABC transporter |
| M. tuberculosis H37Rv | Rv2399c | cysT | 6e-23 | 29.27% (246) | sulfate-transport integral membrane protein ABC transporter |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1889 | modB | 1e-144 | 100.00% (264) | molbdenum-transport integral membrane protein ABC MODB |
| M. gilvum PYR-GCK | Mflv_3860 | - | 3e-95 | 70.87% (254) | NifC-like ABC-type porter |
| M. leprae Br4923 | MLBr_02093 | pstA1 | 3e-09 | 26.58% (222) | membrane-bound component of phosphate transport |
| M. abscessus ATCC 19977 | MAB_2434 | - | 4e-98 | 70.59% (255) | molybdenum ABC transporter permease |
| M. marinum M | MMAR_2732 | modB | 1e-120 | 82.69% (260) | molybdenum-transport integral membrane protein ABC |
| M. avium 104 | MAV_2861 | modB | 1e-119 | 85.83% (254) | molybdate ABC transporter, permease protein |
| M. smegmatis MC2 155 | MSMEG_2015 | modB | 1e-94 | 69.14% (256) | molybdate ABC transporter, permease protein |
| M. thermoresistible (build 8) | TH_3819 | - | 1e-107 | 76.40% (250) | PUTATIVE NifC-like ABC-type porter |
| M. ulcerans Agy99 | MUL_3021 | modB | 1e-118 | 81.92% (260) | molybdenum-transport integral membrane protein ABC |
| M. vanbaalenii PYR-1 | Mvan_2537 | - | 5e-95 | 70.98% (255) | NifC-like ABC-type porter |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3860|M.gilvum_PYR-GCK -MPVRRRAGPIEVGLPEWIYLPAAVG-----------ALFVLTPLAAILL
Mvan_2537|M.vanbaalenii_PYR-1 -MTVRRTGGTIQVGLPVWVYLPAALG-----------ALFVITPLLAVLL
MSMEG_2015|M.smegmatis_MC2_155 MITWRRTRGRVEVGLPAWIFVPATLG-----------ALFVVIPLVAILV
Rv1858|M.tuberculosis_H37Rv --------MHPPTDLPRWVYLPAIAG-----------IVFVAMPLVAIAI
Mb1889|M.bovis_AF2122/97 --------MHPPTDLPRWVYLPAIAG-----------IVFVAMPLVAIAI
MAV_2861|M.avium_104 ------------------MYLPAAAG-----------TMFVVLPLLAIAV
MMAR_2732|M.marinum_M --------MHGPTELPRWVYLPATAG-----------MGLVILPLVAMAA
MUL_3021|M.ulcerans_Agy99 --------MHGPPELPRWVYLPATAG-----------MGLVILPLVAMAA
TH_3819|M.thermoresistible__bu --------MNAST-LPRWIYLPAAVG-----------ATLLALPVFALAG
MAB_2434|M.abscessus_ATCC_1997 --------MTPAPGLPRWVYVPAAIG-----------AALIVLPLLALVV
MLBr_02093|M.leprae_Br4923 -MNLDTLDRPVKAVVLRPISVRRRIKNNVATMFFFTSFLIALVPLVWLLS
: : : *: :
Mflv_3860|M.gilvum_PYR-GCK RIDWPDFVPLITSESSRAALALSLKTAAASTAVCVLLGVPMALVLARGDI
Mvan_2537|M.vanbaalenii_PYR-1 RIDWPNFLPLITSESSRTALLLSLKTATASTVVCVLLGVPMALVLARARF
MSMEG_2015|M.smegmatis_MC2_155 RVEWSQFISLITSESSRAALLLSLKTSAAATAVCTLLGVPMAVVLARARF
Rv1858|M.tuberculosis_H37Rv RVDWPRFWALITTPSSQTALLLSVKTAAASTVLCVLLGVPMALVLARSRG
Mb1889|M.bovis_AF2122/97 RVDWPRFWALITTPSSQTALLLSVKTAAASTVLCVLLGVPMALVLARSRG
MAV_2861|M.avium_104 KVDWPHFWSLITSPSSRTALLLSLRTAAASTALCVALGVPMALVLARGGT
MMAR_2732|M.marinum_M KVDWPNFWSLITSTSSRTALVLSLKTASASTAICVLLGVPMALVLARSRA
MUL_3021|M.ulcerans_Agy99 KVDWPNFWSLITSTSSRTALVLSLKTASASTAICVLLGVPTALVLARSRA
TH_3819|M.thermoresistible__bu RVDWPRFWSLICSESSMTALALSLRTATASTVLCLVLGVPMALALARGGS
MAB_2434|M.abscessus_ATCC_1997 KADWGQFPALVTSPASQAALLLSVRTCLASTALCLLFGVPLAVVLARGPG
MLBr_02093|M.leprae_Br4923 VVVARGFYAVTRFDWWTHSLRGVMPEEFAGGVYHALYGSLVQAVVAAIMA
* .: :* : *. . * .:*
Mflv_3860|M.gilvum_PYR-GCK PGQKVLRALVLLPLVLPPVVGGIALLYTFGR------QGLLGQHLEVLGV
Mvan_2537|M.vanbaalenii_PYR-1 AGQSVLRALVLLPLVLPPVVAGIALLYTFGR------QGLLGQHLEVLGL
MSMEG_2015|M.smegmatis_MC2_155 PGRSVLRALVLLPLVLPPVVGGIALLYTYGR------MGLVGEYLDMAGV
Rv1858|M.tuberculosis_H37Rv RLVRSLRPLILLPLVLPPVVGGIALLYAFGR------LGLIGRYLEAAGI
Mb1889|M.bovis_AF2122/97 RLVRSLRPLILLPLVLPPVVGGIALLYAFGR------LGLIGRYLEAAGI
MAV_2861|M.avium_104 RLVRSLRPLILLPLVLPPVVGGIALLYAFGR------LGLLGHYLEAAGI
MMAR_2732|M.marinum_M RLMRLMRPLILLPLVLPPVVGGIALLYAFGR------LGLIGRYLEAGGI
MUL_3021|M.ulcerans_Agy99 RLMRLMRPLILLPLVLPPVVGGIALLYAFGR------LGLIGRYLEAGGI
TH_3819|M.thermoresistible__bu PILRWIRPLVLLPLVLPPVVGGIALLYAFGR------MGLIGGYLEAVGI
MAB_2434|M.abscessus_ATCC_1997 RMTRLLRPMILLPLVLPPVVGGIALLYAFGK------LGLLGHYLDAAGI
MLBr_02093|M.leprae_Br4923 VPLGSMTAVYLVEYGSGPFVRVTTFMVDVLAGVPSIVAALFIFCLWIATL
: .: *: *.* ::: .*. * :
Mflv_3860|M.gilvum_PYR-GCK RIAFSTTAVVLAQSFVSLPFLVVSLEGALRSAGAGYERIAATLGAAPTTV
Mvan_2537|M.vanbaalenii_PYR-1 RVAFSTTAVVLAQSFVSLPFLVVSLEGALRSAGSGYENVAATLGAKPTTV
MSMEG_2015|M.smegmatis_MC2_155 RIAFTTTAVVLAQSFVSLPFLVVSLEGALRSAGDRYETIAATLGARPTTV
Rv1858|M.tuberculosis_H37Rv SIAFSTAAVVLAQTFVSLPYLVISLEGAARTAGADYEVVAATLGARPGTV
Mb1889|M.bovis_AF2122/97 SIAFSTAAVVLAQTFVSLPYLVISLEGAARTAGADYEVVAATLGARPGTV
MAV_2861|M.avium_104 SVAFSTTAVVLAQTFVSLPFLVISLEGAARTAGADFEVVAATLGARPTTV
MMAR_2732|M.marinum_M TIAFSTTAVVLAQTFVSLPFLVVSLEGAARTAGADYEVVAATLGARPSAV
MUL_3021|M.ulcerans_Agy99 TIAFSTTAVVLAQTFVSLPFLVVPLEGAARTAGADYEVVAATLGARPSAV
TH_3819|M.thermoresistible__bu QIAFSTTAVVLAQTFVSLPFLVLALEGAARTAGTGYEVVAATLGATPTTV
MAB_2434|M.abscessus_ATCC_1997 RIAYSTSAVVLAQTFVSLPFLVISLQGALTTTGQRYDEVAASLGAPPTTV
MLBr_02093|M.leprae_Br4923 GFQQSAFAVSLALVLLMLPVVVRSTEEILRLVPDELREACYALGIPKWKT
. :: ** ** :: ** :* . : . . :** .
Mflv_3860|M.gilvum_PYR-GCK LRTVTLPLVVPGLISGAVLAFARSLGEFGATLTFAGSLQ--------GVT
Mvan_2537|M.vanbaalenii_PYR-1 LRTVTLPLVLPGLVSGAVLAFARSLGEFGATLTFAGSLQ--------GVT
MSMEG_2015|M.smegmatis_MC2_155 LRTVTLPLVLPGILSGAVLAFARSLGEFGATLTFAGSLQ--------GVT
Rv1858|M.tuberculosis_H37Rv WWRVTLPLLLPGVVSGSVLAFARSLGEFGATLTFAGSRQ--------GVT
Mb1889|M.bovis_AF2122/97 WWRVTLPLLLPGVVSGSVLAFARSLGEFGATLTFAGSRQ--------GVT
MAV_2861|M.avium_104 WWRVTLPLLLPGVVSGAVLAFARSLGEFGATLTFAGSRQ--------GVT
MMAR_2732|M.marinum_M WWKVTLPLLMPGLVSGAVLAFARALGEFGATLTFAGSRQ--------GVT
MUL_3021|M.ulcerans_Agy99 WWKVTLPLLMPGLVSGAVLAFARSLGEFGATLTFAGSRQ--------GVT
TH_3819|M.thermoresistible__bu WWRVTLPLLAPGLVSGAVLAFARALGEFGATLTFAGSRQ--------GVT
MAB_2434|M.abscessus_ATCC_1997 LRRVSLPLVMPALASGTVLAFARSLGEFGATLTFAGSRQ--------GVT
MLBr_02093|M.leprae_Br4923 IVRIVFPIATPGIISGVLLSIARVIGETAPVLVLVGYSRSINFDIFDGNM
: :*: *.: ** :*::** :** ...*.:.* : *
Mflv_3860|M.gilvum_PYR-GCK RTLPLEIYLQRETDADAAVALSL-----VLIVVAALIVIGSASR--RLAG
Mvan_2537|M.vanbaalenii_PYR-1 RTLPLEIYLQRETDADAAVALSL-----LLIVLAAGIVIATTGR--RWTG
MSMEG_2015|M.smegmatis_MC2_155 RTLPLEIYLQRETDPDAAVALSL-----VLIAVAAVIVVASASR--KLAG
Rv1858|M.tuberculosis_H37Rv RTLPLEIYLQRVTDPDAAVALSL-----LLVVVAALVVLGVGAR--TPIG
Mb1889|M.bovis_AF2122/97 RTLPLEIYLQRVTDPDAAVALSL-----LLVVVAALVVLGVGAR--TPIG
MAV_2861|M.avium_104 RTLPLEIYLQRVTDADAAVALSI-----LLVAVAAVVVLGLGAR--RLTG
MMAR_2732|M.marinum_M RTLPLEIYLQRVNDPDAAIALSL-----LLVAVAALVVLSLGAR--RLTG
MUL_3021|M.ulcerans_Agy99 RTLPLEIYLQRVNDPDAAIALSL-----LLVAVAALVVLSLGAR--RLTG
TH_3819|M.thermoresistible__bu RTLPLEIYLQREADPEAAVALSM-----VLIAVAAVVVIGLGAR--RLRA
MAB_2434|M.abscessus_ATCC_1997 RTLPLEIYLQRESDPQAAVALSL-----LLVAVAAVVLAVVGARTRRPAG
MLBr_02093|M.leprae_Br4923 ASLPLLIYTELTNPEYAGFLRVWGAALTLIILVAGINLFGAAFRFLATRR
:*** ** : *.. ::: :*. : *
Mflv_3860|M.gilvum_PYR-GCK PL---
Mvan_2537|M.vanbaalenii_PYR-1 SP---
MSMEG_2015|M.smegmatis_MC2_155 PL---
Rv1858|M.tuberculosis_H37Rv TDTR-
Mb1889|M.bovis_AF2122/97 TDTR-
MAV_2861|M.avium_104 TDAR-
MMAR_2732|M.marinum_M GQAG-
MUL_3021|M.ulcerans_Agy99 GQAG-
TH_3819|M.thermoresistible__bu GGWS-
MAB_2434|M.abscessus_ATCC_1997 GGYE-
MLBr_02093|M.leprae_Br4923 CSGCR