For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VPAAPTDRLADTSVPIHPPIAERWSPRAFDPGADLDRDDLVALLEAARWAPTWGRRQPVRFVVGLRGDAP FTTIAGLLRRGNSYAKAASALILLCTDDGEDDRTQRYAAVDGGAAMANITIEAVSRGLIAHPMAGFDVAG ASEAFALPDGLRPVVVIAVGRLGDYADVAPEIAERDRLPRHRLPLSEVVLNWTP
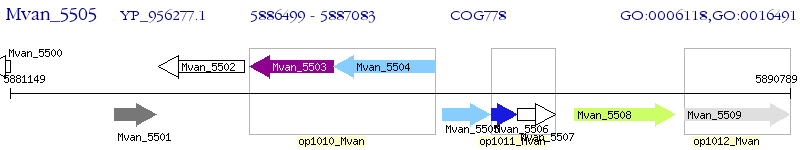
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_5505 | - | - | 100% (194) | nitroreductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_1303 | - | 1e-82 | 75.39% (191) | nitroreductase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0342c | - | 2e-52 | 53.51% (185) | putative nitroreductase family protein |
| M. marinum M | MMAR_0363 | - | 3e-09 | 31.58% (133) | hypothetical protein MMAR_0363 |
| M. avium 104 | MAV_4857 | bluB | 4e-06 | 27.12% (177) | cob(II)yrinic acid a,c-diamide reductase |
| M. smegmatis MC2 155 | MSMEG_6258 | - | 3e-71 | 67.35% (196) | nitroreductase family protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_4756 | - | 7e-09 | 31.82% (132) | hypothetical protein MUL_4756 |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_5505|M.vanbaalenii_PYR-1 MPAAPTDRLADTSVPIHPPIAERWSPRAFDPGADLDRDDLVALLEAARWA
Mflv_1303|M.gilvum_PYR-GCK MPEAPNDRLAATLVPLHPPIARRWSPRAFDPAAELPRDDLIAVLEAARWA
MSMEG_6258|M.smegmatis_MC2_155 MTDAPSDRLANTSVPIHPPLAARWSPRAFDAAAIVTSEQLTAVLEAARWA
MAB_0342c|M.abscessus_ATCC_199 -MGNPTERIANTTVPIQPLIASRWSPRNFDPDADLAPETLTALLEAGRWA
MMAR_0363|M.marinum_M --MGTSADHDHRDLKLWDAFRKRRTHRDFEQ-TELSTEAIGALVEAAASA
MUL_4756|M.ulcerans_Agy99 ---------------MWDAFRKRRTHRDFEQ-TGLSTEAIGALVEAAASA
MAV_4857|M.avium_104 --MTGPAFSPQERRAVYRVISERRDMRRFVAGAVVDEQVLARLLAAAHAA
: : * * * : : : : :: *. *
Mvan_5505|M.vanbaalenii_PYR-1 PTWGRRQPVRFVVGLRG-----------DAPFTTIAGLLRRGNSYAKAAS
Mflv_1303|M.gilvum_PYR-GCK PTWGGRQPVRFVVGVRG-----------DDTFAAIAGLLRRGNRYATAAN
MSMEG_6258|M.smegmatis_MC2_155 ATWGQRQPVRFIVGRRGGQTG---ADAADETFTTLAGLLKRGNSYAHAAS
MAB_0342c|M.abscessus_ATCC_199 ASWGKRFPVRYIVGRRG-----------DATYEALADVLNRGNTYAKVAA
MMAR_0363|M.marinum_M QTFRPG--VRHIVVVTEP-----------ASVATLGDVVPG---WVSNAT
MUL_4756|M.ulcerans_Agy99 QTFRPG--VRHIVVVTEP-----------ASVATLGDVVPG---WVSNAT
MAV_4857|M.avium_104 PSVGLMQPWRFIRITDAALRRRIHALVEQERPRTAAALGERAQEFLALKV
: *.: : . : :
Mvan_5505|M.vanbaalenii_PYR-1 ALILLCTD-------DGEDDRTQ------RYAAVDGGAAMANITIEAVSR
Mflv_1303|M.gilvum_PYR-GCK ALILLCAD-------EGEDDRTR------LYSVVDAGAAMTNLVIEAVAR
MSMEG_6258|M.smegmatis_MC2_155 ALILVCAD-------EGEDERTA------RYAAVDAGAAIAQLTIEAVSR
MAB_0342c|M.abscessus_ATCC_199 ALVLVSTD-------LGDDDNTA------VYATVDAGLAIAQLGVEARSH
MMAR_0363|M.marinum_M AAIVLCTD-------VDKAAALAGSRAVDIYAKLDAGAALAHITLAAVGL
MUL_4756|M.ulcerans_Agy99 AAIVLCTD-------VDKAAALAGSRAVDIYAKLDAGAALAHITLAAVGL
MAV_4857|M.avium_104 EGILECAELLVVALGDDRDKHVFGRRTLPQMDLASVSCAIQNLWLAARAE
:: .:: . . . *: :: : * .
Mvan_5505|M.vanbaalenii_PYR-1 GLIAHPMAGFDVAGASEAFALPDGLRPVVVIAVG-RLGDYADVAPEIAER
Mflv_1303|M.gilvum_PYR-GCK DLIAHPMAGFDVAGAAEAFALPPGLHPLMVVAVG-RLGDYSQVAPEIAER
MSMEG_6258|M.smegmatis_MC2_155 GLIAHPMAGFDVDGARAAFGVPDGVRPFAVVALG-HLGDYATADEAIAER
MAB_0342c|M.abscessus_ATCC_199 GLHTHPMAGFRVEEASSVFGIPGNVRPLAIVAIGPVLDNYDGAEESTVER
MMAR_0363|M.marinum_M GVGICTVGSFTEGAVQAVLDIPDHIRPDVVIGIGIPIRNPLPAPPRAAFQ
MUL_4756|M.ulcerans_Agy99 GVGICTAGSFTEGAVQAVLDIPDHIRPDVVIGIGIPIRNPLPAPLRAAFQ
MAV_4857|M.avium_104 GLGMGWVSLFEPRPLAELLGLPDGAEPVAILCLG-PVPEFPDRPALELDG
.: . * : :* .* :: :* : :
Mvan_5505|M.vanbaalenii_PYR-1 DRLPRHRLPLSEVVLNWTP------
Mflv_1303|M.gilvum_PYR-GCK DSRPRERLPLADVVVNEIVVLNQI-
MSMEG_6258|M.smegmatis_MC2_155 DSRPRERLALEQVAFTGRWG-NPL-
MAB_0342c|M.abscessus_ATCC_199 DHAPRQRPALGDIAFTERWGNSYSG
MMAR_0363|M.marinum_M PGVHYNRFGKPTESTAP--------
MUL_4756|M.ulcerans_Agy99 PGVHYNRFGKPTESTAP--------
MAV_4857|M.avium_104 WATPR---PLAEFVCENRWAQPQLP