For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PEAPNDRLAATLVPLHPPIARRWSPRAFDPAAELPRDDLIAVLEAARWAPTWGGRQPVRFVVGVRGDDTF AAIAGLLRRGNRYATAANALILLCADEGEDDRTRLYSVVDAGAAMTNLVIEAVARDLIAHPMAGFDVAGA AEAFALPPGLHPLMVVAVGRLGDYSQVAPEIAERDSRPRERLPLADVVVNEIVVLNQI*
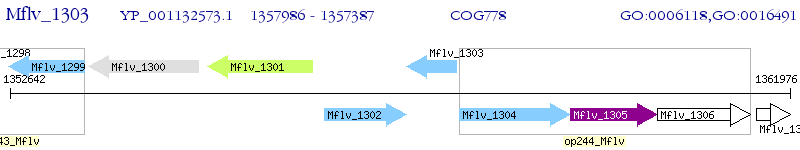
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1303 | - | - | 100% (199) | nitroreductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0342c | - | 3e-47 | 48.68% (189) | putative nitroreductase family protein |
| M. marinum M | MMAR_0555 | - | 5e-06 | 29.07% (172) | oxidoreductase |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_6258 | - | 7e-67 | 64.95% (194) | nitroreductase family protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_1218 | - | 8e-06 | 28.49% (172) | oxidoreductase |
| M. vanbaalenii PYR-1 | Mvan_5505 | - | 1e-82 | 75.39% (191) | nitroreductase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1303|M.gilvum_PYR-GCK MPEAPNDRLAATLVPLHPPIARRWSPRAFDPAAELPRDDLIAVLEAARWA
Mvan_5505|M.vanbaalenii_PYR-1 MPAAPTDRLADTSVPIHPPIAERWSPRAFDPGADLDRDDLVALLEAARWA
MSMEG_6258|M.smegmatis_MC2_155 MTDAPSDRLANTSVPIHPPLAARWSPRAFDAAAIVTSEQLTAVLEAARWA
MAB_0342c|M.abscessus_ATCC_199 -MGNPTERIANTTVPIQPLIASRWSPRNFDPDADLAPETLTALLEAGRWA
MMAR_0555|M.marinum_M --MADYAFTAQERQAVYRVITERRDMRRFVPGAMVPEDVLTRLLHAAHAA
MUL_1218|M.ulcerans_Agy99 --MTDYAFTAQERQAVYRVLTERRDMRHFVPGAVVPEDVLTRLLHAAHAA
* .: :: * . * * . * : : * :*.*.: *
Mflv_1303|M.gilvum_PYR-GCK PTWGGRQPVRFV----VGVRG--------DDTFAAIAGLLRRGNRYATAA
Mvan_5505|M.vanbaalenii_PYR-1 PTWGRRQPVRFV----VGLRG--------DAPFTTIAGLLRRGNSYAKAA
MSMEG_6258|M.smegmatis_MC2_155 ATWGQRQPVRFI----VGRRGGQTGADAADETFTTLAGLLKRGNSYAHAA
MAB_0342c|M.abscessus_ATCC_199 ASWGKRFPVRYI----VGRRG--------DATYEALADVLNRGNTYAKVA
MMAR_0555|M.marinum_M PNVGLMQPWRFIRITDTGLRR-RIHTLVDEERALTAGALGERAEEFLALK
MUL_1218|M.ulcerans_Agy99 PNVGLMQPWRFIRITDTGLRR-RIHTLVDEERALTAGALGERAEEFLALK
.. * * *:: .* * : : . : .*.: :
Mflv_1303|M.gilvum_PYR-GCK NALILLCAD-------EGED------DRTRLYSVVDAGAAMTNLVIEAVA
Mvan_5505|M.vanbaalenii_PYR-1 SALILLCTD-------DGED------DRTQRYAAVDGGAAMANITIEAVS
MSMEG_6258|M.smegmatis_MC2_155 SALILVCAD-------EGED------ERTARYAAVDAGAAIAQLTIEAVS
MAB_0342c|M.abscessus_ATCC_199 AALVLVSTD-------LGDD------DNTAVYATVDAGLAIAQLGVEARS
MMAR_0555|M.marinum_M VEGILECAELMVVALRDGRDGHIFGRRTLPQMDLASVSCAIQNLWLAARA
MUL_1218|M.ulcerans_Agy99 VEGILECAELMVVALRDGRDGHIFGRRTLPQMDLASVSCAIQNLWLAARA
:* .:: * * .. . *: :: : * :
Mflv_1303|M.gilvum_PYR-GCK RDLIAHPMAGFDVAGAAEAFALPPGLHPLMVVAVG-RLGDYSQVAPEIAE
Mvan_5505|M.vanbaalenii_PYR-1 RGLIAHPMAGFDVAGASEAFALPDGLRPVVVIAVG-RLGDYADVAPEIAE
MSMEG_6258|M.smegmatis_MC2_155 RGLIAHPMAGFDVDGARAAFGVPDGVRPFAVVALG-HLGDYATADEAIAE
MAB_0342c|M.abscessus_ATCC_199 HGLHTHPMAGFRVEEASSVFGIPGNVRPLAIVAIGPVLDNYDGAEESTVE
MMAR_0555|M.marinum_M EGLGMGWVSLFDPHRLADLLRMPDGAEPVAVLCLG-PVPEFP--QQPALE
MUL_1218|M.ulcerans_Agy99 EGLGMGWVSLFDPHRLADLLRMPDGAEPVAVLCLG-PVPEFP--QQPALE
..* :: * : :* . .*. ::.:* : :: *
Mflv_1303|M.gilvum_PYR-GCK RDSRPRERLPLADVVVNEIVVLNQI-
Mvan_5505|M.vanbaalenii_PYR-1 RDRLPRHRLPLSEVVLNWTP------
MSMEG_6258|M.smegmatis_MC2_155 RDSRPRERLALEQVAFTGRWG-NPL-
MAB_0342c|M.abscessus_ATCC_199 RDHAPRQRPALGDIAFTERWGNSYSG
MMAR_0555|M.marinum_M LDGWAVAR-PLPEFVSENRWDQP---
MUL_1218|M.ulcerans_Agy99 LDGWAVAR-PLPEFVSENRWDQP---
* . * .* :..