For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MVAQHQPVVTGDAVVLDVQIAQLPVRVLSALIDVTVVFVVYVLGVILWAVTLTQFDPAFSGAVLIIFTVL ALVGYPTIFETATRGKTLGKLALGLRVVSEDGSPERFRQALFRALAGVIELWMLTGGPAVLCSLLSPKGK RIGDIFAGTVVISERAPRQSPPPVMPPQLAWWATTLALSGLSPSQAEIARQFLSRARQLDPQVRDQMAYR IASDVVAQISPPPPPGTPPALVLAAVLAERHRRELGRLQQQPFQPGPVWPAPPQPPAATPPSWPPAPPSA PPPQHPGGFAPPS
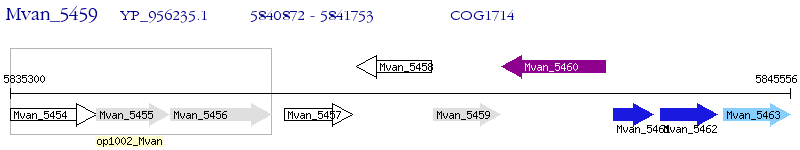
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_5459 | - | - | 100% (293) | RDD domain-containing protein |
| M. vanbaalenii PYR-1 | Mvan_3075 | - | 4e-06 | 35.44% (79) | fibronectin-attachment family protein |
| M. vanbaalenii PYR-1 | Mvan_1068 | - | 6e-06 | 41.54% (65) | glycosyl transferase family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3720 | - | 1e-109 | 62.74% (314) | hypothetical protein Mb3720 |
| M. gilvum PYR-GCK | Mflv_1333 | - | 1e-130 | 79.52% (293) | RDD domain-containing protein |
| M. tuberculosis H37Rv | Rv3695 | - | 1e-109 | 62.74% (314) | hypothetical protein Rv3695 |
| M. leprae Br4923 | MLBr_02312 | - | 7e-65 | 63.54% (181) | hypothetical protein MLBr_02312 |
| M. abscessus ATCC 19977 | MAB_0386c | - | 3e-92 | 59.65% (285) | hypothetical protein MAB_0386c |
| M. marinum M | MMAR_5205 | - | 1e-110 | 66.78% (301) | hypothetical protein MMAR_5205 |
| M. avium 104 | MAV_0408 | - | 1e-113 | 69.76% (291) | integral membrane protein |
| M. smegmatis MC2 155 | MSMEG_6222 | - | 1e-116 | 67.97% (306) | integral membrane protein |
| M. thermoresistible (build 8) | TH_1718 | - | 1e-114 | 67.87% (305) | POSSIBLE CONSERVED MEMBRANE PROTEIN |
| M. ulcerans Agy99 | MUL_4278 | - | 1e-110 | 67.79% (298) | hypothetical protein MUL_4278 |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_5459|M.vanbaalenii_PYR-1 MVAQHQPVVTGDAVVLDVQIAQLPVRVLSALIDVTVVFVVYVLGVILWAV
Mflv_1333|M.gilvum_PYR-GCK MVGQQQPVVTGDAVVLDIQIAQLPVRALSALIDVSVVALVYIIGVILWAM
MSMEG_6222|M.smegmatis_MC2_155 MVSQPEPVVTGDAVVLDVSIAQLPVRVLAILIDIFAIFIFYVVGVLLWSI
TH_1718|M.thermoresistible__bu MAMQPETVVTGDAVVLDVQIAQLPVRAVSALIDAAAIFTLYVLGLMLWAV
Mb3720|M.bovis_AF2122/97 ----MSEVVTGDAVVLDVQIAQLPVRAVSAVIDITIIFIGYILGLMLWAT
Rv3695|M.tuberculosis_H37Rv ----MSEVVTGDAVVLDVQIAQLPVRAVSAVIDITIIFIGYILGLMLWAT
MMAR_5205|M.marinum_M ----MSEVVTGDAVVLDVQIAQLPVRAVSAIIDMTVMFIGYIMGLTLWAA
MUL_4278|M.ulcerans_Agy99 ----MSEVVTGDAVVLDVQIAQLPVRAVSAIIDMTVMFIGYIMGLTLWAA
MAV_0408|M.avium_104 ----MSEVVTGDAVVLDVQIAQLPVRALSALIDIAVIVVGYLLGLMLWAA
MLBr_02312|M.leprae_Br4923 --------------------------------------------------
MAB_0386c|M.abscessus_ATCC_199 ----MSELVTGDAVVLDVKVAQLPVRVVSAFIDIFVQMIVLYGGIFVVAF
Mvan_5459|M.vanbaalenii_PYR-1 TLTQFDPAFSGAVLIIFTVLALVGYPTIFETATRGKTLGKLALGLRVVSE
Mflv_1333|M.gilvum_PYR-GCK TLSQFDPAFSGAVLIIFTVLALVGYPAIFETATRGRTLGKMALGLRVVSD
MSMEG_6222|M.smegmatis_MC2_155 TLTQLDEALSAAILIIFTVLAIVGYPVIMETATRGRSLGKMALGLRVVSE
TH_1718|M.thermoresistible__bu ALASFDDALSAAVLIVFTVLTLVGYPLVFEMATRGRTPGKMAMGLRVVSD
Mb3720|M.bovis_AF2122/97 ALTQFDEALTTAFLIIFTVLALVGYPLVWETATRGRSVGKIVMGLRVVSD
Rv3695|M.tuberculosis_H37Rv ALTQFDEALTTAFLIIFTVLALVGYPLVWETATRGRSVGKIVMGLRVVSD
MMAR_5205|M.marinum_M SLSELDEALTIAFLIIFTVLVLVGYPVIMETATRGRSLGKIAMGLRVVSD
MUL_4278|M.ulcerans_Agy99 SLSELDEALTIAFLIIFTVLVLVGYPVIMETATRGRSLGKIAMGLRVVSD
MAV_0408|M.avium_104 TLTQFDTALSNAILLIFTVLVIVGYPLILETATRGRSVGKIALGLRVVSD
MLBr_02312|M.leprae_Br4923 ------------------------------------------MGLRVVSD
MAB_0386c|M.abscessus_ATCC_199 SLSQFDAALTGALSIVYSVLALVGYPVIWEMATRGRSLGKMAMGLRVVSD
:******:
Mvan_5459|M.vanbaalenii_PYR-1 DGSPERFRQALFRALAGVIELWMLTGGPAVLCSLLSPKGKRIGDIFAGTV
Mflv_1333|M.gilvum_PYR-GCK DGSPERFRQALFRALAGVIEIWMLTGGPAVLCSLVSSKGKRIGDIFAGTV
MSMEG_6222|M.smegmatis_MC2_155 DGSPERFRQALFRGISGFIEIWMLTGGPAVICSLLSSKGKRIGDIFAGTV
TH_1718|M.thermoresistible__bu DGGPERFRQALFRALSSVIEIWMFMGGPAVLCSLISPRGKRIGDYFAGTV
Mb3720|M.bovis_AF2122/97 DGGPERFRQALFRALASVVEIWMLLGSPAVICSMLSPKAKRVGDVFAGTV
Rv3695|M.tuberculosis_H37Rv DGGPERFRQALFRALASVVEIWMLLGSPAVICSMLSPKAKRVGDVFAGTV
MMAR_5205|M.marinum_M DGGPERFRQALFRALAAVVEIWMLLGSPAVICSMLSPKAKRVGDIFAGTI
MUL_4278|M.ulcerans_Agy99 DGGPERFRQALFRALAAVVEIWMLLGSPAVICSMLSPKAKRVGDIFAGTI
MAV_0408|M.avium_104 DGGPERFRQALFRALASLVEIWMLFGSPAVICSILSPKAKRIGDIFAGTV
MLBr_02312|M.leprae_Br4923 DGSPEHFRQALFRVLASVVEIWMLLGSPAVICSMLSPKAKHIGDIFPGTV
MAB_0386c|M.abscessus_ATCC_199 DGGPERLRQAVIRALSAVVEIFMFMGAPAVIASLASTKGKRLGDIFAGTM
**.**::***::* ::..:*::*: *.***:.*: *.:.*::** *.**:
Mvan_5459|M.vanbaalenii_PYR-1 VISERAPRQSPPPVMPPQLAWWATTLALSGLSPSQAEIARQFLSRARQLD
Mflv_1333|M.gilvum_PYR-GCK VISERAPRQAPPPAMPPQLAWWASSLQLSGLAPSQAEMARQFLARAGQLD
MSMEG_6222|M.smegmatis_MC2_155 VISERGPRMQPPPAMPPALAWWASSLQLSGLGPEQAELARQYLNRAQHLD
TH_1718|M.thermoresistible__bu VISERAPRQTPPPPMPPQLAGWAATLELSGLSADQFNLARQYLSRAPQLE
Mb3720|M.bovis_AF2122/97 VVSERGPRLGPPPVMPPSLAWWASSLQLSGLTAGQAEVARQFLVRAPQLD
Rv3695|M.tuberculosis_H37Rv VVSERGPRLGPPPVMPPSLAWWASSLQLSGLTAGQAEVARQFLVRAPQLD
MMAR_5205|M.marinum_M VVSERGPRLAPPPAMPPTLAWWASSLQLSGLSAGQAEVARQFLSRAPQLD
MUL_4278|M.ulcerans_Agy99 VVSERGPRLAPPPAMPPTLAWWASSLQLSGLSAGQAEVARQFLSRAPQLD
MAV_0408|M.avium_104 VVNERGPRLGPPPAMPPSLAWWASSLQLSGLSSGQAEVARQFLSRAAQLD
MLBr_02312|M.leprae_Br4923 VISERRPQRGPSPAMPPSLAWWASSLQLSGLNTRQAELAHQFLSRAPQLD
MAB_0386c|M.abscessus_ATCC_199 VISERGPKLPPPPMMPPQLAQWAQSLQLSGLTSDQADLARQFLNRAGQLA
*:.** *: *.* *** ** ** :* **** . * ::*:*:* ** :*
Mvan_5459|M.vanbaalenii_PYR-1 PQVRDQMAYRIASDVVAQISPPPPPGTPPALVLAAVLAERHRRELGRLQQ
Mflv_1333|M.gilvum_PYR-GCK PHIRDQMAYRVTSDVVAQISPPPPPGVPPALVLAAVLAERHRRELARLQH
MSMEG_6222|M.smegmatis_MC2_155 PRIRDMMGQRILGEVASRISPPPPPGAPPQCVLAAVLAERHRRELARLMP
TH_1718|M.thermoresistible__bu PAVRDEMAYRIAADLAPRISPPPPPGTPPHHLLAAVVAERHRRELARLRP
Mb3720|M.bovis_AF2122/97 PALREQMAYRIAGDVVARIAPPPPPGVPPQLVLAAVLAERHRRELLRLRP
Rv3695|M.tuberculosis_H37Rv PALREQMAYRIAGDVVARIAPPPPPGVPPQLVLAAVLAERHRRELLRLRP
MMAR_5205|M.marinum_M LRLRHQMAYRIAGDVLSHIAPPPPPGTPPQLVLAAVLAERHRRELARLR-
MUL_4278|M.ulcerans_Agy99 LRLRHQMAYRIAGDVLSHIAPPPPPGTPPQLVLAAVLAERHRRELARLR-
MAV_0408|M.avium_104 PGLRLQMAYRIAGDVVARIAPPPPPGAPPELVLAAVLAERHRRELARLR-
MLBr_02312|M.leprae_Br4923 PQLQEQMAYRITGDVMSRIAPPPPSSTPPQLMLAAVLAERHRHKLVQLR-
MAB_0386c|M.abscessus_ATCC_199 PHTRDQMLYQISTDVLASIAPPPPPGTPPQFMLAAVLAERHRRALAQMR-
: * :: :: . *:****...** :****:*****: * ::
Mvan_5459|M.vanbaalenii_PYR-1 QPFQPG-------------PVWPAPPQPPAATPP-------SWPPAPPSA
Mflv_1333|M.gilvum_PYR-GCK P--APG-------------PGWSAAP------------------PTSPPP
MSMEG_6222|M.smegmatis_MC2_155 TPPPPPSPP--------TTPPWSPPGYPAAGYPPQPPVPAYSPPPAVPAH
TH_1718|M.thermoresistible__bu QAAVPAA------------PVWYPPQQPGHPVYPAPPYQPQYQPPQQPAQ
Mb3720|M.bovis_AF2122/97 TLPPAGQAPWAQMAPHRGWPPGLSGATPWSPQQPV-IPWPEPDPPPQAAP
Rv3695|M.tuberculosis_H37Rv TLPPAGQAPWAQMAPHRGWPPGLSGATPWSPQQPV-IPWPEPDPPPQAAP
MMAR_5205|M.marinum_M ---PAGAG-----APP-GWPPQTPPAPVWWPGQPAPQPQIQPQFAPDPAP
MUL_4278|M.ulcerans_Agy99 ---PAGAG-----APP-GWPPQTPPAPVWWPGQPAPQPLIQPPFAPDPAP
MAV_0408|M.avium_104 -------------------PPAPWPAPGYPPAWPGSGPAPQWPAPGPANP
MLBr_02312|M.leprae_Br4923 -------------------PTPVGPT----PSSPTVSKYPVPPRVAWAGI
MAB_0386c|M.abscessus_ATCC_199 -------------------PDYPATPP------------PAPSAPPPAPE
* .
Mvan_5459|M.vanbaalenii_PYR-1 PPPQHP--GGFAPPS
Mflv_1333|M.gilvum_PYR-GCK PPPAAG--GGFTPPS
MSMEG_6222|M.smegmatis_MC2_155 SPPQQPSAGGFAPPR
TH_1718|M.thermoresistible__bu QPPDPSP-GGFVPPS
Mb3720|M.bovis_AF2122/97 WPQQAPDGPGFSPPG
Rv3695|M.tuberculosis_H37Rv WPQQAPDGPGFSPPG
MMAR_5205|M.marinum_M SPPQGPVGGGFSPPS
MUL_4278|M.ulcerans_Agy99 SPPQGPVGGGFSPPS
MAV_0408|M.avium_104 GPPEG-FSAGFTPPR
MLBr_02312|M.leprae_Br4923 CSTQFLVASGHR---
MAB_0386c|M.abscessus_ATCC_199 PSESSTGRSGFATPG
. *.