For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
NSSQWLDISILALALIAAISGWRSGAPGSLLALVGVALGAAAGILLAPHVVSHISGPRTKLFVTLFLILA LVVIGEIAGVVLGRAVRGAIRSPGLRVADSVVGVALQLVLVLTAAWLLGTALVSSPQPNLAAAVQGSKVI AEVDSVAPSWLRSVPNRLSALWETAGLHDVLKPFGPTPVAEIDPPDPSLATSPVVNTTRSSVVKVRGVAQ SCQKVLEGTGFVVAPNRVMSNAHVVAGSDSVTVEVDGQTYDAGVVSYDPNADISILDVPNLPSAPLQFVD AEAQPGTDGIVMGYPGGGEFTATPARVREVIELNGPDIYRSTTVTREVYTIRGTVKQGNSGGPMINRQGR VLGVVFGAAVDDVDTGFVLTADEVARQMAKVGNVERVATGTCISAG*
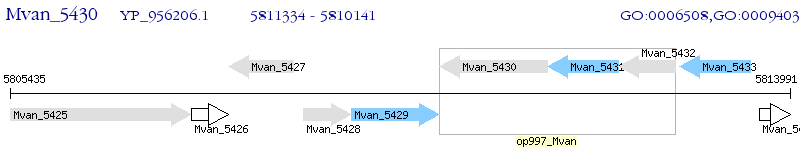
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_5430 | - | - | 100% (397) | colicin V production protein |
| M. vanbaalenii PYR-1 | Mvan_5537 | - | 1e-13 | 31.71% (205) | PDZ/DHR/GLGF domain-containing protein |
| M. vanbaalenii PYR-1 | Mvan_4494 | - | 5e-10 | 27.78% (288) | peptidase S1 and S6, chymotrypsin/Hap |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3695c | - | 1e-141 | 63.48% (397) | putative membrane-associated serine protease |
| M. gilvum PYR-GCK | Mflv_1376 | - | 0.0 | 87.91% (397) | colicin V production protein |
| M. tuberculosis H37Rv | Rv3671c | - | 1e-141 | 63.22% (397) | membrane-associated serine protease |
| M. leprae Br4923 | MLBr_02298 | - | 1e-144 | 65.08% (398) | putative membrane-associated serine protease |
| M. abscessus ATCC 19977 | MAB_0421 | - | 1e-150 | 66.67% (396) | membrane-associated serine protease |
| M. marinum M | MMAR_5159 | - | 1e-146 | 65.49% (397) | membrane-associated serine protease |
| M. avium 104 | MAV_0459 | - | 1e-150 | 67.51% (397) | serine protease |
| M. smegmatis MC2 155 | MSMEG_6183 | - | 1e-171 | 75.25% (396) | serine protease |
| M. thermoresistible (build 8) | TH_0962 | - | 1e-157 | 69.77% (397) | serine protease |
| M. ulcerans Agy99 | MUL_4245 | - | 1e-146 | 65.24% (397) | membrane-associated serine protease |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_5430|M.vanbaalenii_PYR-1 -----MNSSQWLDISILALALIAAISGWRSGAPGSLLALVGVALGAAAGI
Mflv_1376|M.gilvum_PYR-GCK -----MNSSQWLDIAILAVAMLAAVSGWRSGAPGSLMALVGVVLGAGAGI
MSMEG_6183|M.smegmatis_MC2_155 -----MTPSLWLDLAVVAIAFVAAISGWRSGALGSLMSFIGVVLGAVAGV
TH_0962|M.thermoresistible__bu -----MTAAQWLDFAILAVALIAAISGWRSGAVGSLMSFLGVVLGAVAGV
MAB_0421|M.abscessus_ATCC_1997 ---MNLNPSQWLDIGVIAVAFIAAVSGWRSGALGSLMSFVGVILGAVAGI
Mb3695c|M.bovis_AF2122/97 -----MTPSQWLDIAVLAVAFIAAISGWRAGALGSMLSFGGVLLGATAGV
Rv3671c|M.tuberculosis_H37Rv -----MTPSQWLDIAVLAVAFIAAISGWRAGALGSMLSFGGVLLGATAGV
MMAR_5159|M.marinum_M --MNSMTPSQWLDIAVLAVAFIAAVSGWRSGALGSMLSFAGVLLGAIAGV
MUL_4245|M.ulcerans_Agy99 --MNSMTPSQWLDIAVLAVAFIAAVSGWRSGALGSMLSFAGVLLGAIAGV
MAV_0459|M.avium_104 MNLNSMTPSQWLDVAVLAVAFIAAISGWRSGALGSLLSFVGVLLGAIAGV
MLBr_02298|M.leprae_Br4923 --MSSMTLSEWLDIAVLAVAFIAAISGWRSGALGSLLSLIGVLFGAMAGV
:. : ***..::*:*::**:****:** **:::: ** :** **:
Mvan_5430|M.vanbaalenii_PYR-1 LLAPHVVSHISGPRTKLFVTLFLILALVVIGEIAGVVLGRAVRGAIRSPG
Mflv_1376|M.gilvum_PYR-GCK LLAPHVVDHISGPRTKLFATLFLILVLVVIGEIAGVVLGRAVRGAIRNPG
MSMEG_6183|M.smegmatis_MC2_155 LLAPHVIPHVDGARTKLFVTLFLILALVVVGEIAGVVLGRAVRGTIRSGL
TH_0962|M.thermoresistible__bu LLAPHVVTYIDGARTRLFATLFLILALVVIGEIAGVVLGRAARSALRSGP
MAB_0421|M.abscessus_ATCC_1997 MLAPHVVANIEGSRTKLFASLLLILVLVVIGEVAGVVLGRAMRGAIRNRV
Mb3695c|M.bovis_AF2122/97 LLAPHIVSQISAPRAKLFAALFLILALVVVGEVAGVVLGRAVRGAIRNRP
Rv3671c|M.tuberculosis_H37Rv LLAPHIVSQISAPRAKLFAALFLILALVVVGEVAGVVLGRAVRGAIRNRP
MMAR_5159|M.marinum_M LLAPHIVSHINAPRAKLFAALFLILGLVVVGEVAGVVLGRAVRGAIRNRP
MUL_4245|M.ulcerans_Agy99 LLAPHIVSHINAPRAKLFAALFLILGLVVVGEVAGVVLGRAVRGAIRNRP
MAV_0459|M.avium_104 LLAPHLVSHIAAPRAKLFAALFLILALVVIGEVAGVVLGRAVRGAIRSRS
MLBr_02298|M.leprae_Br4923 LLVPHVVSHISAPRAKLFAALFLILALVVIGEVVGVVLGRAVRSANRSRT
:*.**:: : ..*::**.:*:*** ***:**:.******* *.: *.
Mvan_5430|M.vanbaalenii_PYR-1 LRVADSVVGVALQLVLVLTAAWLLGTALVSSP-QPNLAAAVQGSKVIAEV
Mflv_1376|M.gilvum_PYR-GCK LRIVDSVVGVALQLVLVLTAAWLLGTALVSSP-QPNLAAAVSGSRVIAEV
MSMEG_6183|M.smegmatis_MC2_155 FRAFDSVVGVSLMLVAVLVAAWLLGS-LLTSSDQPNLAAAVKGSRVLSEV
TH_0962|M.thermoresistible__bu VRALDSVTGVAMQLVAVLLAAWLLATPLASTSGQPAVAAAVSDSRLLSEV
MAB_0421|M.abscessus_ATCC_1997 LRTGDSVVGVVLQVAAVLVAAWLLSIPMQSSN-QPNISAAARESKVLSQV
Mb3695c|M.bovis_AF2122/97 IRLIDSVIGVGVQLVVVLTAAWLLAMPLTQSKEQPELAAAVKGSRVLARV
Rv3671c|M.tuberculosis_H37Rv IRLIDSVIGVGVQLVVVLTAAWLLAMPLTQSKEQPELAAAVKGSRVLARV
MMAR_5159|M.marinum_M IRVIDSVIGVGVQLVVVLTAAWLLATPLTQSKDQPELAAAVRGSRVLAQV
MUL_4245|M.ulcerans_Agy99 IRVIDSVIGVGVQLVVVLTAAWLLATPLTQSKDQPELAAAVRGSRVLAQV
MAV_0459|M.avium_104 IRTVDSVIGVGVQLVVVLTAAWLLATPLTQSKDQPELAAAVRGSRVLAKV
MLBr_02298|M.leprae_Br4923 VRLVDSVIGVGVQLIVVLTAAWLLATPLTQSKEQPELASAVRGSRALASV
.* *** ** : : ** *****. : : ** :::*. *: :: *
Mvan_5430|M.vanbaalenii_PYR-1 DSVAPSWLRSVPNRLSALWETAGLHDVLKPFGPTPVAEIDPPDPSLATSP
Mflv_1376|M.gilvum_PYR-GCK DAVAPDWLRSVPDRMSSVWENAGLHDVLKPFGPTPVAAVDAPDPALATSP
MSMEG_6183|M.smegmatis_MC2_155 DRVAPTWLRQVPTRLSGLLDTSGLPDVLEPFGRTPIVNVDAPDAALADDA
TH_0962|M.thermoresistible__bu DKVAPEWLRSVPQRISALLDDSGLPDVLEPFGRTPIAAVDEPDPALAQDT
MAB_0421|M.abscessus_ATCC_1997 DKYAPDQLRKVPNDLSKLLDTSGLPSVLQPFGKTPIAAVDAPDATLADGP
Mb3695c|M.bovis_AF2122/97 NEAAPTWLKTVPKRLSALLNTSGLPAVLEPFSRTPVIPVASPDPALVNNP
Rv3671c|M.tuberculosis_H37Rv NEAAPTWLKTVPKRLSALLNTSGLPAVLEPFSRTPVIPVASPDPALVNNP
MMAR_5159|M.marinum_M NEVAPTWLKTVPKRLSGLLNTSGLPAVLEPFSRTPVIPVASPDPALARNP
MUL_4245|M.ulcerans_Agy99 NAVAPTWLKTVPKRLSGLLNTSGLPAVLEPFSRTPVIPVASPDPALARNP
MAV_0459|M.avium_104 NEVAPPWLKTVPKRLSALLNTSGLPAVLEPFSRTPVIPVASPDPELASNP
MLBr_02298|M.leprae_Br4923 NEVAPPWLKTVPKRLSALLNTSGLPAVLEPFSRTPVLPVASPDPALVDNP
: ** *: ** :* : : :** **:**. **: : **. *. ..
Mvan_5430|M.vanbaalenii_PYR-1 VVNTTRSSVVKVRGVAQSCQKVLEGTGFVVAPNRVMSNAHVVAGSDSVTV
Mflv_1376|M.gilvum_PYR-GCK VVNQTRSSVVKVKGVAQSCQKVLEGTGFVVAPNRVMSNAHVVAGSDTVTV
MSMEG_6183|M.smegmatis_MC2_155 VVAATRPSVVKIRGVAPSCQKVLEGSGFVVAPSRVMSNAHVVAGADSVTV
TH_0962|M.thermoresistible__bu VVGLTAPSVVKIRGVAEVCRKVLEGSGFVIAPNRVMSNAHVVAGADQVTI
MAB_0421|M.abscessus_ATCC_1997 VVRGSEPSVVKIKGIARSCQKSLEGTGFVIAPHRVMSNAHVVAGTNSVTV
Mb3695c|M.bovis_AF2122/97 VVAATEPSVVKIRSLAPRCQKVLEGTGFVISPDRVMTNAHVVAGSNNVTV
Rv3671c|M.tuberculosis_H37Rv VVAATEPSVVKIRSLAPRCQKVLEGTGFVISPDRVMTNAHVVAGSNNVTV
MMAR_5159|M.marinum_M VVEATAPSVLKIRSLAPSCQKVLEGSGFVIAPDRVMTNAHVVAGANSVQV
MUL_4245|M.ulcerans_Agy99 VVEATAPSVLKIRSLAPSCQKVLEGSGFVIAPDRVMTNAHVVAGANSVQV
MAV_0459|M.avium_104 VVQATAPSVVKVRSLAPSCQKVLEGSGFVIAPDRVMTNAHVVAGSNSVQI
MLBr_02298|M.leprae_Br4923 VVLATQPSVVKIRSVAPSCQKVLEGTGFVVSLDRVMTNAHVVAGSDGVQV
** : .**:*::.:* *:* ***:***:: ***:*******:: * :
Mvan_5430|M.vanbaalenii_PYR-1 EVDGQTYDAGVVSYDPNADISILDVPNLPSAPLQFVDAEAQPGTDGIVMG
Mflv_1376|M.gilvum_PYR-GCK EVDGQEYTAGVVSYDPNADISILDVPDLPSVPLQFVDALAQTGTDGIVLG
MSMEG_6183|M.smegmatis_MC2_155 EVDGETYDAHVVSYDPDADISILDVPNLPSAPLAFAADPAPQGTDAVVMG
TH_0962|M.thermoresistible__bu DADGQEYPATVVSYDPDIDISILDVPDLPLRPLQFVDHEAPPGTDAVAMG
MAB_0421|M.abscessus_ATCC_1997 ESAGQTYDASVVSYDPNADISILAVPDMPAAPLVFASKPAKTGDDAIVLG
Mb3695c|M.bovis_AF2122/97 YAGDKPFEATVVSYDPSVDVAILAVPHLPPPPLVFAAEPAKTGADVVVLG
Rv3671c|M.tuberculosis_H37Rv YAGDKPFEATVVSYDPSVDVAILAVPHLPPPPLVFAAEPAKTGADVVVLG
MMAR_5159|M.marinum_M FASGKPLDATVVSYDPSVDIAILAVPGLPPPPLIFAEREAPTGTNVVVLG
MUL_4245|M.ulcerans_Agy99 FASGKPLDATVVSYDPSVDIAILAVPGLPPPPLIFAEREAPTGTNVVVLG
MAV_0459|M.avium_104 YASGNPLDATVVSYDPSVDIAILAVPNLPPPPLPFAQTEAKTGASVVVLG
MLBr_02298|M.leprae_Br4923 YASGNPFDATVVSYDPSVDIAILAVPHLPPAPLRFDDTKAKPGTNVVMLG
.: * ******. *::** ** :* ** * * * . : :*
Mvan_5430|M.vanbaalenii_PYR-1 YPGGGEFTATPARVREVIELNGPDIYRS-TTVTREVYTIRGTVKQGNSGG
Mflv_1376|M.gilvum_PYR-GCK YPGGGDFTATPARVREVIELNGPDIYRS-TTVTREVYTIRGTVRQGNSGG
MSMEG_6183|M.smegmatis_MC2_155 YPGGGDFLATPARVREIIELNGPDIYRT-TTVTREVYTVRGTVRQGNSGG
TH_0962|M.thermoresistible__bu YPGGGDFVATPVRVRETITLKGPDIYQS-KTVTRSVYTVRGSIKQGNSGG
MAB_0421|M.abscessus_ATCC_1997 YPGGGDYKASPARVREIIELKGPDIYRS-ITVTREVYTVRGSVLQGNSGG
Mb3695c|M.bovis_AF2122/97 YPGGGNFTATPARIREAIRLSGPDIYGDPEPVTRDVYTIRADVEQGDSGG
Rv3671c|M.tuberculosis_H37Rv YPGGGNFTATPARIREAIRLSGPDIYGDPEPVTRDVYTIRADVEQGDSGG
MMAR_5159|M.marinum_M YPGGGNFTATPARIREAIRLSGPDIYRNPQPVTRDVYTIRANVEQGDSGG
MUL_4245|M.ulcerans_Agy99 YPGGGNFTATPARIREATRLSGPDIYRNLQPVTRDVYTIRANVEQGDSGG
MAV_0459|M.avium_104 YPGGGNFTATPARIRELIKLSGPDIYRDPAPVTRDVYTIRASVEQGNSGG
MLBr_02298|M.leprae_Br4923 YPGGGNFTATSARIRQAIKLTGPDIYRGLAPVTRDVYTIRATVEQGNSGG
*****:: *:..*:*: *.***** .***.***:*. : **:***
Mvan_5430|M.vanbaalenii_PYR-1 PMINRQGRVLGVVFGAAVDDVDTGFVLTADEVARQMAKVGNVER-VATGT
Mflv_1376|M.gilvum_PYR-GCK PMVNRQGKVLGVVFGAAVDDIDTGFVLTADEVAAQMARVGNVDL-VPTGT
MSMEG_6183|M.smegmatis_MC2_155 PMINRAGKVLGVVFGAAVDDVDTGFVLTAKEVERQLAKIGNTER-VATGT
TH_0962|M.thermoresistible__bu PMIDRSGKVLGVVFGAAVDDDDTGFVLTAKQVEHQIARVGNTEP-VPTGQ
MAB_0421|M.abscessus_ATCC_1997 PLIDTEGRVLGVVFGAAIDDDDTGFALTAKQVKDQLAKADLADP-VATGS
Mb3695c|M.bovis_AF2122/97 PLIDLNGQVLGVVFGAAVDDAETGFVLTAGEVAGQLAKIGATQP-VGTGA
Rv3671c|M.tuberculosis_H37Rv PLIDLNGQVLGVVFGAAIDDAETGFVLTAGEVAGQLAKIGATQP-VGTGA
MMAR_5159|M.marinum_M PLIDLNGQVLGVVFGAAVDDPDTGFVLTSGEVASQLARIGATER-VATGS
MUL_4245|M.ulcerans_Agy99 PLIDLNGQVLGVVFGAAVDDPDTGFVLTSGEVASQLARIGATER-VATGS
MAV_0459|M.avium_104 PLIDLNGQVLGVVFGAAVDDPDTGFVLTADEVAGQLAKIGETKT-VGTGS
MLBr_02298|M.leprae_Br4923 PLIDLNGRVLGVVFGAAVDDSETGFVLTAEEVSHRLTEIGETQEAVATGA
*::: *:*********:** :***.**: :* :::. . .. * **
Mvan_5430|M.vanbaalenii_PYR-1 CISAG----------------------
Mflv_1376|M.gilvum_PYR-GCK CISSG----------------------
MSMEG_6183|M.smegmatis_MC2_155 CINS-----------------------
TH_0962|M.thermoresistible__bu CVGAPAP--------------------
MAB_0421|M.abscessus_ATCC_1997 CVSAAEANRPTEQPVNGPQSPVPGKAP
Mb3695c|M.bovis_AF2122/97 CVS------------------------
Rv3671c|M.tuberculosis_H37Rv CVS------------------------
MMAR_5159|M.marinum_M CVS------------------------
MUL_4245|M.ulcerans_Agy99 CVS------------------------
MAV_0459|M.avium_104 CVS------------------------
MLBr_02298|M.leprae_Br4923 CVS------------------------
*:.