For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MHALVDPFQADMVLRALVAGVIAACLCSLVGCWVLLRGNVFLGEAMTHGMLPGVALAALLGISLLIGGLV AALVMAVGVAALGRSSRLSSDTSVGLLLVGMLALGVIVVSRSQSFAVDLTGFLFGDVFAVRTADLVVLAT ALVLTLGAVIIGHRAFVAVTFDPRKAATLGLRPQLAIPVLTMLMAVAMVASFHVVGTLLVLGLLVAPPAA ALAWSRSIPRVMLLSALIGSAAVYLGLLISWYAGTAGGATIAGVAVLIFFASTLLSFGRGNWKRMSVVAA SACVVAGCGTGGESPAPAEHPTSAGEDEVVVEGARELDGALTRLVLVDPATGATSVYDAVDEVETSIGSY GPVDGISGDGRFAYLRTGGRTTVVDAGAWTFGHGDHDHYFATEPAEVATLDVPAASVSAVNSIVAVRTPT NATALFDREKLADNVVEAPAGLALSPDVAAVAPYGSRLVTVTAAGRMQVTGDTGTTDLVGECPNPSWASP TRRAVVFGCDTGAVRVTGGDGDLAVTPAPFPADAPAERPRQMEHRDRADVFAGVSAGTVWVLDSRQRSWT LIPVADAVATNTAGDGTVLVLRRDGTLSAYEVNARAETARIPLFPEGIPADGAQPVVDLDSDRAYVNNAA TREVYEIDYADGLRIARTLRTEVAPGLMVEAGR*
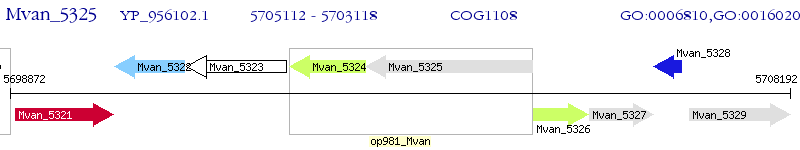
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_5325 | - | - | 100% (664) | ABC-3 protein |
| M. vanbaalenii PYR-1 | Mvan_3369 | - | 3e-36 | 29.59% (267) | ABC-3 protein |
| M. vanbaalenii PYR-1 | Mvan_5318 | - | 6e-19 | 28.00% (225) | ABC-3 protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2086 | - | 5e-07 | 26.36% (129) | integral membrane protein |
| M. gilvum PYR-GCK | Mflv_3166 | - | 2e-38 | 32.58% (267) | ABC-3 protein |
| M. tuberculosis H37Rv | Rv2060 | - | 5e-07 | 26.36% (129) | integral membrane protein |
| M. leprae Br4923 | MLBr_00335 | - | 1e-18 | 25.10% (255) | putative ABC-transporter transmembrane protein |
| M. abscessus ATCC 19977 | MAB_0575c | - | 8e-19 | 25.10% (255) | ABC transporter transmembrane protein |
| M. marinum M | MMAR_5073 | - | 2e-21 | 27.45% (255) | permease component of an ABC-type transporter |
| M. avium 104 | MAV_0581 | - | 1e-21 | 27.45% (255) | ABC-type Mn2+/Zn2+ transport system, permease component |
| M. smegmatis MC2 155 | MSMEG_6045 | - | 1e-22 | 26.53% (245) | ABC heavy metal transporter, inner membrane subunit |
| M. thermoresistible (build 8) | TH_4073 | - | 4e-38 | 32.03% (256) | PUTATIVE ABC-3 protein |
| M. ulcerans Agy99 | MUL_4149 | - | 2e-21 | 27.45% (255) | permease component of an ABC-type transporter |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_5073|M.marinum_M MTNDRLVDMLDHLFAFDITAHLLRHDFVQQALIAAALLGLVAGLIGPFVV
MUL_4149|M.ulcerans_Agy99 MTNDRLVDILDHLFAFDITAHLLRHDFVQQALIAAALLGLVAGLIGPFVV
Mb2086|M.bovis_AF2122/97 --------------------------------------------------
Rv2060|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00335|M.leprae_Br4923 -MDDRLAHMWNHLFSFDITTHLLSHNFVQQALLAALLLGLVAGLIGPFIV
MAV_0581|M.avium_104 -MEHRLADMADHLFSFDITVHLLGHDFVQQALVAAALLGLVAGLIGPFIV
MAB_0575c|M.abscessus_ATCC_199 ---------MNNFFDLSLTADLLGRDFVQQAILAAALLGLLAGLIGPFVV
MSMEG_6045|M.smegmatis_MC2_155 MPPGDLAQTYLAIGYQHNWLHILGSAFMRNALIGGTLVALAAGLIGYFII
Mflv_3166|M.gilvum_PYR-GCK -----------MTVV-EFLVEPLQLEFMVRATVTTLIASTVCATLSCWLV
TH_4073|M.thermoresistible__bu -----------MTLLREILIDPLAYEFMVRALATALITSVVCALLSCWLV
Mvan_5325|M.vanbaalenii_PYR-1 --------------MMHALVDPFQADMVLRALVAGVIAACLCSLVGCWVL
MMAR_5073|M.marinum_M MRQMSFAVHGSSELSLTGAAFALLSGLPVGVGALVGSALAAALFGILGQR
MUL_4149|M.ulcerans_Agy99 MRQMSFAVHGSSELSLTGAAFALLSGLPVGVGALVGSALAAALFGILGQR
Mb2086|M.bovis_AF2122/97 --------------------------------------------------
Rv2060|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00335|M.leprae_Br4923 MRQMSFAVHGSSELSLTGATFALLAGFEMGVGALVGSALAATLFGILGRR
MAV_0581|M.avium_104 MRQMSFAVHGSSELSLTGAAFALLVGIGVGVGALIGSALAAALFGVLGRR
MAB_0575c|M.abscessus_ATCC_199 MRQMSFAVHGSSELSLTGAAAALLAGANVGTGALLGSVVAAILFGVLGQR
MSMEG_6045|M.smegmatis_MC2_155 VRNTAFAAHALAHIGLPGATGAALLGLPAVLGLGVFCVAGAVVIGALGKQ
Mflv_3166|M.gilvum_PYR-GCK LIGWSLMGDAVSHAVLPGVVLAHIVGAPFALGAIIFGFVAVGLIGVVRDT
TH_4073|M.thermoresistible__bu LIGWSLMGDAVSHAVLPGVVLAYIVGAPFAVGAVVFGFLAVALIGLVRDT
Mvan_5325|M.vanbaalenii_PYR-1 LRGNVFLGEAMTHGMLPGVALAALLGISLLIGGLVAALVMAVGVAALGRS
MMAR_5073|M.marinum_M ARERD-SVIGVVLAFGMGLAVLFIHLYPGRTGTSFALLTGQIVGVGYSGL
MUL_4149|M.ulcerans_Agy99 ARERD-SVIGVVLAFGMGLAVLFIHLYPGRTGTSFALLTGQIVGVGYSGL
Mb2086|M.bovis_AF2122/97 --------------------------------------------------
Rv2060|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00335|M.leprae_Br4923 ARERD-SVIGVVLAFGLGLAVLFIHLYPGRTGTSLALLTGQIIDVGYSGL
MAV_0581|M.avium_104 ARERD-SVIGVVLAFGLGLAVLFIHLYPGRTATSFALLTGQIVGVGYTGL
MAB_0575c|M.abscessus_ATCC_199 TKDRD-SSIGVVMAFGLGLAVLFIHLYPGRTGTNFALLTGQIVGVGYSGL
MSMEG_6045|M.smegmatis_MC2_155 AHDRE-VATGTVLALATGFGLFFNSLATKNSSTLTNVLFGNLLAITHEQL
Mflv_3166|M.gilvum_PYR-GCK SRTKEDAAIGVVFTTLFALGLVLISVTPSQVDLN-HIIFGNVLGVSRADL
TH_4073|M.thermoresistible__bu SRVKEDAAIGIVFTTLFAAGLVLVSVTPSQVDLN-HIVFGNLLGVSRNDL
Mvan_5325|M.vanbaalenii_PYR-1 SRLSSDTSVGLLLVGMLALGVIVVSRSQSFAVDLTGFLFGDVFAVRTADL
MMAR_5073|M.marinum_M TMLALICVLVVAALAICYRPLLFATVDPEVAAARGVPVRTLSIVFAALVG
MUL_4149|M.ulcerans_Agy99 TMLALICVLVVAALAICYRPLLFATVDPEVAAARGVPVRTLSIVFVALVG
Mb2086|M.bovis_AF2122/97 -MLTVVCLLVVTVLAICYRPLLFATVDPEVAAARGVPVRALGIVFAALMG
Rv2060|M.tuberculosis_H37Rv -MLTVVCLLVVTVLAICYRPLLFATVDPEVAAARGVPVRALGIVFAALMG
MLBr_00335|M.leprae_Br4923 ATLALVCLFVIAVLTTCYRPLLFATVDPEVAAACGVPVHALGIVFAALVG
MAV_0581|M.avium_104 TMLALVCLLVIAVLATCYRPLLFATVDPDVAAARGVPVHALGIVFAALVG
MAB_0575c|M.abscessus_ATCC_199 LMLVVVSMVVGAVLAVTYRPMLFATVDPDVAEARGVPVRALGIVFAALVG
MSMEG_6045|M.smegmatis_MC2_155 LTFTALLLVLAAAIVFIYRPLLFASVNAQVAEAKGVPVRTLSVVFMVLLG
Mflv_3166|M.gilvum_PYR-GCK IQVAVLGTVVMAVLVYKRRDFTLYAFDPHHAHAIGLNPRLLGAALLGLLA
TH_4073|M.thermoresistible__bu LQVAILGAVVVAAVLAKRRDFTLYAFDSIHAHTIGLNPRLLGAALLALLA
Mvan_5325|M.vanbaalenii_PYR-1 VVLATALVLTLGAVIIGHRAFVAVTFDPRKAATLGLRPQLAIPVLTMLMA
.. . .: * : :.:. * : *: : .: *:.
MMAR_5073|M.marinum_M VVAAQAVQIVGALLVMSLLITPAAAAARVVTAPAAAILTSVVFAEVAAVG
MUL_4149|M.ulcerans_Agy99 VVAAQAVQIVGALLVMSLLITPAAAAARVVTAPAAAILTSVVFAELAAVG
Mb2086|M.bovis_AF2122/97 VVAAQAVQIVGALLVMSLLITPAAAAARVVVAPVAAIATSVVFAEVSAVG
Rv2060|M.tuberculosis_H37Rv VVAAQAVQIVGALLVMSLLITPAAAAARVVVAPVAAIATSVVFAEVSAVG
MLBr_00335|M.leprae_Br4923 LVAAQAVQVVGALLVMSLLITPAAAAAQVVASPVAAMVAAVIFAEVSAVG
MAV_0581|M.avium_104 VVAAQAVQIVGALLVMSLLITPAAAAARVVASPGAAMLASVAFAEVSALG
MAB_0575c|M.abscessus_ATCC_199 LTAAQGVQIVGALLVMSLLITPAAAAARITSSPARAIALSIVFAEIAAIG
MSMEG_6045|M.smegmatis_MC2_155 VAVTMAVQAVGTLLLFALVVTPAATAITLTAQPRLAMAISTVISLLSVWA
Mflv_3166|M.gilvum_PYR-GCK LTAVVALQAVGVVLVVALLITPGATAYLITERFSRMLLIAPTIAAVCALT
TH_4073|M.thermoresistible__bu LTAVAALQAVGVVLVVAMLIIPGATAYLLTDSFGRMLVIAPLMSVAAAIT
Mvan_5325|M.vanbaalenii_PYR-1 VAMVASFHVVGTLLVLGLLVAPPAAALAWSRSIPRVMLLSALIGSAAVYL
:. . ..: **.:*:..::: * *:* : : :. ..
MMAR_5073|M.marinum_M GIVLSLAPGVPVSVFVATISFLIYLICRLLG--------------RHR--
MUL_4149|M.ulcerans_Agy99 GIVLSLAPGVPVSVFVATISFLIYLICRLLG--------------RHR--
Mb2086|M.bovis_AF2122/97 GILLSLAPGVPVSVFVATISFVIYLICWLLR--------------RRR--
Rv2060|M.tuberculosis_H37Rv GILLSLAPGVPVSVFVATISFVIYLICWLLR--------------RRR--
MLBr_00335|M.leprae_Br4923 GVVLSLAPGVPVSVFVATISFLIYLFCWLLG--------------RHREV
MAV_0581|M.avium_104 GIVLSLAPGVPVSVFVATISFLIYLACWLIG--------------RRREA
MAB_0575c|M.abscessus_ATCC_199 GIMLSLAPGVPISVFVTMIAVAIYLLCRLVG--------------RYRHA
MSMEG_6045|M.smegmatis_MC2_155 GLVLSAVFNLPPSFVIVTIACVFWFVVWAAN--------------RARRS
Mflv_3166|M.gilvum_PYR-GCK GLYASYYLDTASGPMVVLANGTAFALAYLAR-----LGREQ--RTRHHR-
TH_4073|M.thermoresistible__bu GLYVSYHLDTASGGMIVVAQGLLFGLVYLFAPRQGVLTRAASARLRSRRS
Mvan_5325|M.vanbaalenii_PYR-1 GLLISWYAGTAGGATIAGVAVLIFFASTLLSFGR---------GNWKRMS
*: * . . . :. : :
MMAR_5073|M.marinum_M --------------------------------------------------
MUL_4149|M.ulcerans_Agy99 --------------------------------------------------
Mb2086|M.bovis_AF2122/97 --------------------------------------------------
Rv2060|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00335|M.leprae_Br4923 CA------------------------------------------------
MAV_0581|M.avium_104 AT------------------------------------------------
MAB_0575c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_6045|M.smegmatis_MC2_155 AVARP---------------------------------------------
Mflv_3166|M.gilvum_PYR-GCK PGVR-----APWTETPAAEDASDCRRSRR--AGLPRR---CGRRP-----
TH_4073|M.thermoresistible__bu PGDREHAEIANLGTTGSMETCSRRSVAARGPGGAPTSSTSAGSRPGRAAW
Mvan_5325|M.vanbaalenii_PYR-1 VVAASACVVAGCGTGGESPAPAEHPTSAGEDEVVVEGARELDGALTRLVL
MMAR_5073|M.marinum_M --------------------------------------------------
MUL_4149|M.ulcerans_Agy99 --------------------------------------------------
Mb2086|M.bovis_AF2122/97 --------------------------------------------------
Rv2060|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00335|M.leprae_Br4923 --------------------------------------------------
MAV_0581|M.avium_104 --------------------------------------------------
MAB_0575c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_6045|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_3166|M.gilvum_PYR-GCK --------------------------------------------------
TH_4073|M.thermoresistible__bu WTPRRRSPGWCRSSNGCATPIRTS--------------------------
Mvan_5325|M.vanbaalenii_PYR-1 VDPATGATSVYDAVDEVETSIGSYGPVDGISGDGRFAYLRTGGRTTVVDA
MMAR_5073|M.marinum_M --------------------------------------------------
MUL_4149|M.ulcerans_Agy99 --------------------------------------------------
Mb2086|M.bovis_AF2122/97 --------------------------------------------------
Rv2060|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00335|M.leprae_Br4923 --------------------------------------------------
MAV_0581|M.avium_104 --------------------------------------------------
MAB_0575c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_6045|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_3166|M.gilvum_PYR-GCK --------------------------------------------------
TH_4073|M.thermoresistible__bu --------------------------------------------------
Mvan_5325|M.vanbaalenii_PYR-1 GAWTFGHGDHDHYFATEPAEVATLDVPAASVSAVNSIVAVRTPTNATALF
MMAR_5073|M.marinum_M --------------------------------------------------
MUL_4149|M.ulcerans_Agy99 --------------------------------------------------
Mb2086|M.bovis_AF2122/97 --------------------------------------------------
Rv2060|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00335|M.leprae_Br4923 --------------------------------------------------
MAV_0581|M.avium_104 --------------------------------------------------
MAB_0575c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_6045|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_3166|M.gilvum_PYR-GCK --------------------------------------------------
TH_4073|M.thermoresistible__bu --------------------------------------------------
Mvan_5325|M.vanbaalenii_PYR-1 DREKLADNVVEAPAGLALSPDVAAVAPYGSRLVTVTAAGRMQVTGDTGTT
MMAR_5073|M.marinum_M --------------------------------------------------
MUL_4149|M.ulcerans_Agy99 --------------------------------------------------
Mb2086|M.bovis_AF2122/97 --------------------------------------------------
Rv2060|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00335|M.leprae_Br4923 --------------------------------------------------
MAV_0581|M.avium_104 --------------------------------------------------
MAB_0575c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_6045|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_3166|M.gilvum_PYR-GCK --------------------------------------------------
TH_4073|M.thermoresistible__bu --------------------------------------------------
Mvan_5325|M.vanbaalenii_PYR-1 DLVGECPNPSWASPTRRAVVFGCDTGAVRVTGGDGDLAVTPAPFPADAPA
MMAR_5073|M.marinum_M --------------------------------------------------
MUL_4149|M.ulcerans_Agy99 --------------------------------------------------
Mb2086|M.bovis_AF2122/97 --------------------------------------------------
Rv2060|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00335|M.leprae_Br4923 --------------------------------------------------
MAV_0581|M.avium_104 --------------------------------------------------
MAB_0575c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_6045|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_3166|M.gilvum_PYR-GCK --------------------------------------------------
TH_4073|M.thermoresistible__bu --------------------------------------------------
Mvan_5325|M.vanbaalenii_PYR-1 ERPRQMEHRDRADVFAGVSAGTVWVLDSRQRSWTLIPVADAVATNTAGDG
MMAR_5073|M.marinum_M --------------------------------------------------
MUL_4149|M.ulcerans_Agy99 --------------------------------------------------
Mb2086|M.bovis_AF2122/97 --------------------------------------------------
Rv2060|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00335|M.leprae_Br4923 --------------------------------------------------
MAV_0581|M.avium_104 --------------------------------------------------
MAB_0575c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_6045|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_3166|M.gilvum_PYR-GCK --------------------------------------------------
TH_4073|M.thermoresistible__bu --------------------------------------------------
Mvan_5325|M.vanbaalenii_PYR-1 TVLVLRRDGTLSAYEVNARAETARIPLFPEGIPADGAQPVVDLDSDRAYV
MMAR_5073|M.marinum_M -------------------------------------
MUL_4149|M.ulcerans_Agy99 -------------------------------------
Mb2086|M.bovis_AF2122/97 -------------------------------------
Rv2060|M.tuberculosis_H37Rv -------------------------------------
MLBr_00335|M.leprae_Br4923 -------------------------------------
MAV_0581|M.avium_104 -------------------------------------
MAB_0575c|M.abscessus_ATCC_199 -------------------------------------
MSMEG_6045|M.smegmatis_MC2_155 -------------------------------------
Mflv_3166|M.gilvum_PYR-GCK -------------------------------------
TH_4073|M.thermoresistible__bu -------------------------------------
Mvan_5325|M.vanbaalenii_PYR-1 NNAATREVYEIDYADGLRIARTLRTEVAPGLMVEAGR