For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SAPALEVGTKLPELKIYGDPTFIVSTAIATRDYQDVHHDRDKAQAKGSKDIFVNILTDTGLVQRYLTDWA GPTARIKSIGLRLGVPWYAYDTITFTGEVTAVDDGVATVKVLGSNSLGDHVIATATLSLGDR*
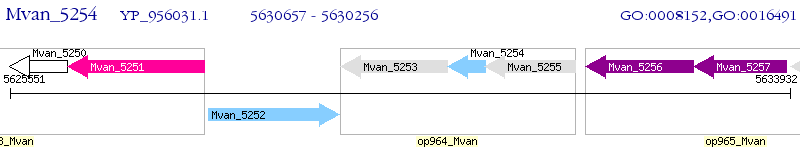
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_5254 | - | - | 100% (133) | MaoC-like dehydratase |
| M. vanbaalenii PYR-1 | Mvan_3988 | - | 9e-06 | 27.97% (118) | dehydratase |
| M. vanbaalenii PYR-1 | Mvan_5844 | - | 4e-05 | 23.08% (130) | dehydratase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3571c | - | 1e-57 | 81.60% (125) | hypothetical protein Mb3571c |
| M. gilvum PYR-GCK | Mflv_1512 | - | 3e-67 | 90.91% (132) | dehydratase |
| M. tuberculosis H37Rv | Rv3541c | - | 1e-57 | 81.60% (125) | hypothetical protein Rv3541c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0617 | - | 3e-52 | 75.00% (124) | hypothetical protein MAB_0617 |
| M. marinum M | MMAR_5028 | - | 4e-62 | 81.68% (131) | hypothetical protein MMAR_5028 |
| M. avium 104 | MAV_0620 | - | 8e-64 | 83.33% (132) | hypothetical protein MAV_0620 |
| M. smegmatis MC2 155 | MSMEG_5991 | - | 1e-63 | 86.36% (132) | MaoC like domain-containing protein |
| M. thermoresistible (build 8) | TH_0365 | - | 4e-63 | 84.09% (132) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_4102 | - | 3e-61 | 83.33% (126) | hypothetical protein MUL_4102 |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_5254|M.vanbaalenii_PYR-1 ---MSAPALEVGTKLPELKIYGDPTFIVSTAIATRDYQDVHHDRDKAQAK
Mflv_1512|M.gilvum_PYR-GCK ---MSAPSIEVGTTLPELTIYGDPTFIVSTAIATRDYQDVHHDRDKAQAK
MSMEG_5991|M.smegmatis_MC2_155 MGAQSMTVIEVGTALPELAIYGDPTFIVSTAIATRDYQDVHHDRDKAQAK
TH_0365|M.thermoresistible__bu ---MSTPVVEVGTKLPELKIHADPTFIVSTALATRDFQDVHHDRDKAQAK
MMAR_5028|M.marinum_M ---MSAPTMDVGTKLPELKLHGDPTFIVSTALATRDFQDVHHDRDKAQAK
MUL_4102|M.ulcerans_Agy99 --------MDVGTKLPELKLHGDPTFIVSTALATRDFQDVHHDRDKAQAK
Mb3571c|M.bovis_AF2122/97 -------MTVVGAVLPELKLYGDPTFIVSTALATRDFQDVHHDRDKAVAQ
Rv3541c|M.tuberculosis_H37Rv -------MTVVGAVLPELKLYGDPTFIVSTALATRDFQDVHHDRDKAVAQ
MAV_0620|M.avium_104 ---MSAPVIEVGTTLPELKLYGDPTFIISTALATRDFQDVHHDRDKAQAK
MAB_0617|M.abscessus_ATCC_1997 -------MTTVGEKLPELKLEGTPTFIVSTALATRDFQDVHHDRDLAQAK
** **** : . ****:***:****:******** * *:
Mvan_5254|M.vanbaalenii_PYR-1 GSKDIFVNILTDTGLVQRYLTDWAGPTARIKSIGLRLGVPWYAYDTITFT
Mflv_1512|M.gilvum_PYR-GCK GSKDIFVNILTDTGLVQRYLTDWAGPAARIRSIGLRLGVPWYAYDTITFT
MSMEG_5991|M.smegmatis_MC2_155 GSKDIFVNILTDTGLVQRYLTDWAGPNARIKSIGLRLGVPWYAYDTVTFR
TH_0365|M.thermoresistible__bu GSKDIFVNILTDTGLVQRYLTDWAGPAARIKSIKLRLGVPWYAYDTVTFT
MMAR_5028|M.marinum_M GSKDIFVNILTDTGLVQRYVTDWAGPSALIKSIGLRLGVPWYAYDTVIFS
MUL_4102|M.ulcerans_Agy99 GSKDIFVNILTDTGLVQRYVTDWAGPSALIKSIGLRLGLPWYAYDTVTFS
Mb3571c|M.bovis_AF2122/97 GSKDIFVNILTDTGLVQRYVTDWAGPSALIKSIGLRLGVPWYAYDTVTFS
Rv3541c|M.tuberculosis_H37Rv GSKDIFVNILTDTGLVQRYVTDWAGPSALIKSIGLRLGVPWYAYDTVTFS
MAV_0620|M.avium_104 GSKDIFVNILTDTGLVQRYVTDWAGPSALIKSIGLRLGVPWYAYDTVTFS
MAB_0617|M.abscessus_ATCC_1997 GSKDIFINILSDTGLVERFVTDWAGPAARVKSISLRLGVPWYAYDTTVFT
******:***:*****:*::****** * ::** ****:******* *
Mvan_5254|M.vanbaalenii_PYR-1 GEVTAVDDGVATVKVLGSNSLGDHVIATATLSLGDR-
Mflv_1512|M.gilvum_PYR-GCK GEVTAVDDDIATVKVVGANSLGNHVIATATLALGDH-
MSMEG_5991|M.smegmatis_MC2_155 GEVTAIEDDLVTVKVTGSNSLGNHVIATATLNLGDKA
TH_0365|M.thermoresistible__bu GEVTDVVDGLVTVKVVGSNSLGDHIIATTTLTMGES-
MMAR_5028|M.marinum_M GEVTTIEDGLITVKVVGSNSLGNHVIATVTLTVGGA-
MUL_4102|M.ulcerans_Agy99 GEVTAIEDGLITVKVVGSNSLGNHVIATVTLTVGGA-
Mb3571c|M.bovis_AF2122/97 GEVTAVNDGLITVKVVGRNTLGDHVTATVELSMRDS-
Rv3541c|M.tuberculosis_H37Rv GEVTAVNDGLITVKVVGRNTLGDHVTATVELSMRDS-
MAV_0620|M.avium_104 GEVTAVQDGLVTLKVFGRNSLGDHVIATVTLTMGDA-
MAB_0617|M.abscessus_ATCC_1997 GEVAAVEDGVVEVDVVGNNSLGAHVTAKVKLTIGAEQ
***: : *.: :.* * *:** *: *.. * :