For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VLRFDPFSDLDALTRGLLTSQTGSSRSPRFMPMDLCKIDDHYVLTADLPGVDPGSVDVNVDNGTLTISAH RTARSEESTQWLANERFFGSYRRQLSLGDGVDTAAISATYENGVLTVTIPVAEKAKPRKIEISHTGNQKS IQPTTVEAG
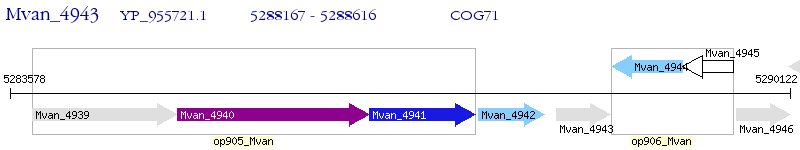
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_4943 | - | - | 100% (149) | heat shock protein Hsp20 |
| M. vanbaalenii PYR-1 | Mvan_3941 | - | 2e-13 | 38.04% (92) | heat shock protein Hsp20 |
| M. vanbaalenii PYR-1 | Mvan_1374 | - | 2e-13 | 39.13% (92) | heat shock protein Hsp20 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2057c | hspX | 5e-12 | 40.40% (99) | heat shock protein hspX |
| M. gilvum PYR-GCK | Mflv_1805 | - | 9e-71 | 84.46% (148) | heat shock protein Hsp20 |
| M. tuberculosis H37Rv | Rv2031c | hspX | 6e-12 | 40.40% (99) | heat shock protein hspX |
| M. leprae Br4923 | MLBr_01795 | hsp18 | 8e-24 | 39.42% (137) | 18 KDA antigen (HSP 16.7) |
| M. abscessus ATCC 19977 | MAB_3467c | - | 3e-25 | 40.85% (142) | heat shock protein |
| M. marinum M | MMAR_2991 | - | 2e-27 | 42.48% (153) | molecular chaperone (small heat shock protein) |
| M. avium 104 | MAV_4106 | - | 3e-27 | 41.50% (147) | Hsp20/alpha crystallin family protein |
| M. smegmatis MC2 155 | MSMEG_5611 | - | 7e-69 | 86.52% (141) | spore protein |
| M. thermoresistible (build 8) | TH_0596 | - | 1e-64 | 76.35% (148) | spore protein |
| M. ulcerans Agy99 | MUL_2232 | - | 1e-27 | 42.48% (153) | molecular chaperone (small heat shock protein) |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_4943|M.vanbaalenii_PYR-1 -----------MLRFDPFSDLDALTRGLLTSQTGSSRSP----RFMPMDL
Mflv_1805|M.gilvum_PYR-GCK -----------MLRFDPFSDIDALTRGLLTSQTGSNRTP----RFMPMDL
MSMEG_5611|M.smegmatis_MC2_155 -----------MLRFDPFSDLDALTKGLLSTGTGSSRTP----RFMPMDL
TH_0596|M.thermoresistible__bu VAGLGSQEVIAVLRFDPFSDLDEWARSMLSTQVGSNRAP----RFMPMDL
MMAR_2991|M.marinum_M ----------MLMRTDPFRDLDRFAQQVL----GTSARP----AVMPMDA
MUL_2232|M.ulcerans_Agy99 ----------MLMRTDPFRDLDRFAQQVL----GTSARP----AVMPMDA
MLBr_01795|M.leprae_Br4923 ----------MLMRTDPFRELDRFAEQVL----GTSARP----AVMPMDA
MAV_4106|M.avium_104 ----------MLMRSDPFRELDRLTNQVL----GTATRP----AVMPMDA
MAB_3467c|M.abscessus_ATCC_199 ----------MLMRTDPFRDLDRFTQQVL----GTAARP----ALMPMDA
Mb2057c|M.bovis_AF2122/97 ------MATTLPVQRHPRSLFPEFSELFAAFPSFAGLRPTFDTRLMRLED
Rv2031c|M.tuberculosis_H37Rv ------MATTLPVQRHPRSLFPEFSELFAAFPSFAGLRPTFDTRLMRLED
:: .* : :. . : * .* ::
Mvan_4943|M.vanbaalenii_PYR-1 CKIDDHYVLTADLPGVDPG-SVDVNVDNGTLTISAHRTARSEESTQWLAN
Mflv_1805|M.gilvum_PYR-GCK CKIDDHYVLTADLPGVDPG-SVDVDVDNGTLTISAHRTARSDESAQWLSS
MSMEG_5611|M.smegmatis_MC2_155 CKIDDHYVLTADLPGVDPG-SVDVTVDNGMLTISAHRTARSDESAQWLAN
TH_0596|M.thermoresistible__bu CKIDDHYVLTADLPGVDPG-SVDVSVENGMLTISAHRSARSDESAQWLAN
MMAR_2991|M.marinum_M WREGDKFVVEFDLPGIDAD-SLDIDIERNVVTVRAERPA-VDPNREMLAS
MUL_2232|M.ulcerans_Agy99 WREGDKFVVEFDLPGIDAD-SLDIDIERNVVTVRAERPA-VDPNREMLAS
MLBr_01795|M.leprae_Br4923 WREGEEFVVEFDLPGIKAD-SLDIDIERNVVTVRAERPG-VDPDREMLAA
MAV_4106|M.avium_104 WREGEHFVVEFDLPGIDAD-SLDIDIERNVLTVRAERPA-LDPNREMLAS
MAB_3467c|M.abscessus_ATCC_199 WRDGNEFIVEFDLPGISGE-SLDLDIEHNVVTVRAERAP-VDPGREMLAS
Mb2057c|M.bovis_AF2122/97 EMKEGRYEVRAELPGVDPDKDVDIMVRDGQLTIKAERTEQKDFDG---RS
Rv2031c|M.tuberculosis_H37Rv EMKEGRYEVRAELPGVDPDKDVDIMVRDGQLTIKAERTEQKDFDG---RS
.: : :***:. .:*: : . :*: *.*. : .
Mvan_4943|M.vanbaalenii_PYR-1 ERFFGSYRRQLSLGDGVDTAAISATYENGVLTVTIPVAEKAKPRKIEISH
Mflv_1805|M.gilvum_PYR-GCK ERFYGTYRRQLSLGDGIDTGAISATYENGVLTVTIPVAAKARPRKIAINH
MSMEG_5611|M.smegmatis_MC2_155 ERFFGSYRRQLSLGEGIDTSAIAATYENGVLTVTIPMVERAKPRKIEIAH
TH_0596|M.thermoresistible__bu ERFFGNFRRQISLGEGVDTSAIKATYENGVLTVTIPLAESAKPRKIAVER
MMAR_2991|M.marinum_M ERPRGVFSRQLVLGENLDTARIAASYTEGVLKLQIPVAEKAKPRKISITR
MUL_2232|M.ulcerans_Agy99 ERPRGVFSRQLVLGENLDTARIAASYTEGVLKLQIPVAEKAKPRKISITR
MLBr_01795|M.leprae_Br4923 ERPRGVFNRQLVLGENLDTERILASYQEGVLKLSIPVAERAKPRKISVDR
MAV_4106|M.avium_104 ERPRGVFSRELVLGDNLDTDKIEASYRDGVLSLHIPVAEKAKPRKIAVGR
MAB_3467c|M.abscessus_ATCC_199 ERARGVFSRQLVLGDNLDTEKIAADYRNGVLRLRIPVAEKAKPRKIVVDR
Mb2057c|M.bovis_AF2122/97 EFAYGSFVRTVSLPVGADEDDIKATYDKGILTVSVAVSE-GKPTEKHIQI
Rv2031c|M.tuberculosis_H37Rv EFAYGSFVRTVSLPVGADEDDIKATYDKGILTVSVAVSE-GKPTEKHIQI
* * : * : * . * * * * .*:* : :.: .:* : :
Mvan_4943|M.vanbaalenii_PYR-1 T----GNQKSIQPT--TVEAG-
Mflv_1805|M.gilvum_PYR-GCK T----KSQTSIEAN--TVDAE-
MSMEG_5611|M.smegmatis_MC2_155 G----GEQKSITVEGETVSNDQ
TH_0596|M.thermoresistible__bu A----GEQKTIEAA--TVDAE-
MMAR_2991|M.marinum_M G-AGDKTISENGAHPEVIEA--
MUL_2232|M.ulcerans_Agy99 G-AGDKTISENVAHPEVIEA--
MLBr_01795|M.leprae_Br4923 GNNGHQTIN-KTAH-EIIDA--
MAV_4106|M.avium_104 G-EAPRAVT-ETAR-EVVNA--
MAB_3467c|M.abscessus_ATCC_199 PADAVVIEGENDSARGKISA--
Mb2057c|M.bovis_AF2122/97 R----------STN--------
Rv2031c|M.tuberculosis_H37Rv R----------STN--------