For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ATTLPVQRHPRSLFPEFSELFAAFPSFAGLRPTFDTRLMRLEDEMKEGRYEVRAELPGVDPDKDVDIMVR DGQLTIKAERTEQKDFDGRSEFAYGSFVRTVSLPVGADEDDIKATYDKGILTVSVAVSEGKPTEKHIQIR STN*
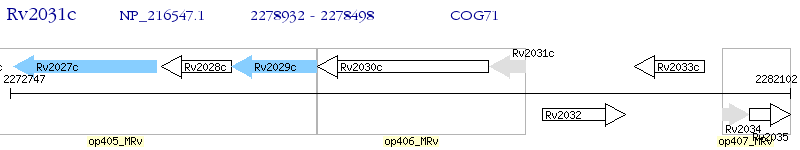
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv2031c | hspX | - | 100% (144) | HEAT SHOCK PROTEIN HSPX (ALPHA-CRSTALLIN HOMOLOG) (14 kDa ANTIGEN) (HSP16.3) |
| M. tuberculosis H37Rv | Rv0251c | hsp | 2e-12 | 41.84% (98) | heat shock protein hsp (heat-stress-induced ribosome-binding |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2057c | hspX | 1e-79 | 100.00% (144) | heat shock protein hspX |
| M. gilvum PYR-GCK | Mflv_1805 | - | 4e-12 | 41.41% (99) | heat shock protein Hsp20 |
| M. leprae Br4923 | MLBr_01795 | hsp18 | 8e-05 | 29.13% (103) | 18 KDA antigen (HSP 16.7) |
| M. abscessus ATCC 19977 | MAB_4402 | - | 7e-13 | 38.06% (134) | heat shock protein Hsp20 |
| M. marinum M | MMAR_3484 | hspX_1 | 8e-57 | 72.34% (141) | heat shock protein HspX_1 |
| M. avium 104 | MAV_4906 | - | 2e-13 | 43.48% (92) | Hsp20/alpha crystallin family protein |
| M. smegmatis MC2 155 | MSMEG_3932 | - | 2e-46 | 60.84% (143) | 14 kDa antigen |
| M. thermoresistible (build 8) | TH_1239 | hspX | 2e-46 | 62.14% (140) | HEAT SHOCK PROTEIN HSPX (ALPHA-CRSTALLIN HOMOLOG) (14 kDa |
| M. ulcerans Agy99 | MUL_1178 | hsp | 3e-10 | 41.98% (81) | heat shock protein Hsp |
| M. vanbaalenii PYR-1 | Mvan_1374 | - | 2e-46 | 60.87% (138) | heat shock protein Hsp20 |
CLUSTAL 2.0.9 multiple sequence alignment
Rv2031c|M.tuberculosis_H37Rv ---MATTLPVQRHPRSLFPEFSELFAAFPSFAGLRPTFDTRLMRLEDEMK
Mb2057c|M.bovis_AF2122/97 ---MATTLPVQRHPRSLFPEFSELFAAFPSFAGLRPTFDTRLMRLEDEMK
MMAR_3484|M.marinum_M ---MG-TLSVQRPARSLFPEFSELFAAFPSFAGLRPAFDSRLMKLEDEMK
MSMEG_3932|M.smegmatis_MC2_155 ---MT-KLPERSRARSLFPEMSDFFAGLPSWASIRPVFGDHIIRIEDEMK
Mvan_1374|M.vanbaalenii_PYR-1 ---MN-NVAVQQPRRSAWPEVADFFAGFPSWANLRPMFGGHLIKVEDDIK
TH_1239|M.thermoresistible__bu MKEMIMTTVERQRAKSLFPEIADFFAGFPSWPSLRPMFDTNLMRLEDELT
MAV_4906|M.avium_104 --MSNLALWTR-PAWDTDRWLRDFFGPAAAADWNKPATSAFKPAAEIVKD
MUL_1178|M.ulcerans_Agy99 --MSNLALWSR-PAWSTDRWLRDFFGPAAASDWYKPLATGFNPAAEIVKD
MAB_4402|M.abscessus_ATCC_1997 --MSNLVWTRRSPFAEFDALVRQSFGPATS--WPTP---GFVPSADVVKD
Mflv_1805|M.gilvum_PYR-GCK --------MLRFDPFSDIDALTRGLLTSQTGSNRTP----RFMPMDLCKI
MLBr_01795|M.leprae_Br4923 -------MLMRTDPFRELDRFAEQVLG----TSARP----AVMPMDAWRE
: . . * :
Rv2031c|M.tuberculosis_H37Rv EGRYEVRAELPGVDPDKDVDIMVRDGQLTIKAERTEQKDFDG-------R
Mb2057c|M.bovis_AF2122/97 EGRYEVRAELPGVDPDKDVDIMVRDGQLTIKAERTEQKDFDG-------R
MMAR_3484|M.marinum_M GDRYELRAEIPGVDPEKDIEITVLDGQLTIKAERSEKKEFNG-------R
MSMEG_3932|M.smegmatis_MC2_155 DGGYELRAEVPGIDPAKDIDITVRDGVLTIKAERTEKKETNG-------R
Mvan_1374|M.vanbaalenii_PYR-1 DGKYELQAEIPGVDPEKDVDITVRDGVLTIKAERSEKKESNG-------R
TH_1239|M.thermoresistible__bu DDKYMVRAEIPGVDPDKDIEITVQDGRLTIRAERQEKKETNG-------R
MAV_4906|M.avium_104 GDDAIVRVELPGVDVDQDVQVELDRGRLVIRGEHRDEHAEEK---DGRTL
MUL_1178|M.ulcerans_Agy99 GDDAVVRLELPGVDVENDVNVEVERGELVIRGEHRDEHAQEAGEANRRTL
MAB_4402|M.abscessus_ATCC_1997 GDDALVRLDLPGVDVGKDVTVEVERGTLVVSGERRDERAEQT---EGRTL
Mflv_1805|M.gilvum_PYR-GCK DDHYVLTADLPGVDPG-SVDVDVDNGTLTISAHRTARSDESA----QWLS
MLBr_01795|M.leprae_Br4923 GEEFVVEFDLPGIKAD-SLDIDIERNVVTVRAERPG-VDPDR----EMLA
: ::**:. .: : : . :.: ..: .
Rv2031c|M.tuberculosis_H37Rv SEFAYGSFVRTVSLPVGADEDDIKATYDKGILTVSVAVSEGK-PTEKHIQ
Mb2057c|M.bovis_AF2122/97 SEFAYGSFVRTVSLPVGADEDDIKATYDKGILTVSVAVSEGK-PTEKHIQ
MMAR_3484|M.marinum_M SEFCYGSFIRTVPLPAGVTEDGIEASYDKGILTVSVPVAEAK-PAERRVR
MSMEG_3932|M.smegmatis_MC2_155 SEFAYGSFMRSVTLPPGADEDAIKAGYDKGILTISVPVTEPEAAAEKRVA
Mvan_1374|M.vanbaalenii_PYR-1 SEFSYGSFTRAVTLPAGADEDGIKASYDKGILSVSVPLKESE-APEKHIA
TH_1239|M.thermoresistible__bu SEFNYGSFVRTVSLPASADEENITADYDKGILTVSVPLRETQ-PSPKRIE
MAV_4906|M.avium_104 REIRYGSFHRSFQLPGHVTDDDITASYDAGVLTVRVTGAY---AGNQAKR
MUL_1178|M.ulcerans_Agy99 REIRYGSFRRSFQLPEHVTSEAVSASYDAGVLTVRVQGAYRDPAASEPQR
MAB_4402|M.abscessus_ATCC_1997 REVRYGTFRRTFTLPTHVTGDAISASYDAGILTVKVAGAY---ANAAAQK
Mflv_1805|M.gilvum_PYR-GCK SERFYGTYRRQLSLGDGIDTGAISATYENGVLTVTIPVAAKARPRKIAIN
MLBr_01795|M.leprae_Br4923 AERPRGVFNRQLVLGENLDTERILASYQEGVLKLSIPVAERAKPRKISVD
* * : * . * : * *: *:*.: : .
Rv2031c|M.tuberculosis_H37Rv IRSTN--------------
Mb2057c|M.bovis_AF2122/97 IRSTN--------------
MMAR_3484|M.marinum_M VQSVN--------------
MSMEG_3932|M.smegmatis_MC2_155 VETVSP-------------
Mvan_1374|M.vanbaalenii_PYR-1 VEAAN--------------
TH_1239|M.thermoresistible__bu VKAGT--------------
MAV_4906|M.avium_104 IAITK--------------
MUL_1178|M.ulcerans_Agy99 IAITK--------------
MAB_4402|M.abscessus_ATCC_1997 IEITT--------------
Mflv_1805|M.gilvum_PYR-GCK HTKSQTSIEANTVDAE---
MLBr_01795|M.leprae_Br4923 RGNNGHQTINKTAHEIIDA