For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MLMRTDPFRDLDRFAQQVLGTSARPAVMPMDAWREGDKFVVEFDLPGIDADSLDIDIERNVVTVRAERPA VDPNREMLASERPRGVFSRQLVLGENLDTARIAASYTEGVLKLQIPVAEKAKPRKISITRGAGDKTISEN GAHPEVIEA
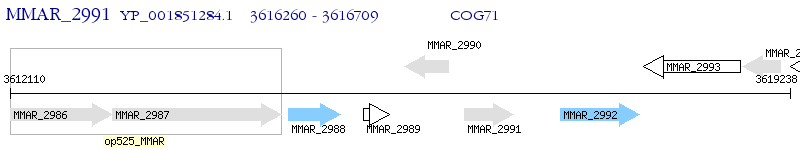
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_2991 | - | - | 100% (149) | molecular chaperone (small heat shock protein) |
| M. marinum M | MMAR_0515 | hsp | 4e-08 | 29.27% (123) | heat shock protein Hsp |
| M. marinum M | MMAR_3484 | hspX_1 | 3e-06 | 31.43% (105) | heat shock protein HspX_1 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0257c | hsp | 2e-05 | 31.86% (113) | heat shock protein hsp (heat-stress-induced ribosome-binding |
| M. gilvum PYR-GCK | Mflv_1805 | - | 1e-26 | 40.27% (149) | heat shock protein Hsp20 |
| M. tuberculosis H37Rv | Rv0251c | hsp | 2e-05 | 31.86% (113) | heat shock protein hsp (heat-stress-induced ribosome-binding |
| M. leprae Br4923 | MLBr_01795 | hsp18 | 4e-64 | 82.00% (150) | 18 KDA antigen (HSP 16.7) |
| M. abscessus ATCC 19977 | MAB_3467c | - | 1e-56 | 78.79% (132) | heat shock protein |
| M. avium 104 | MAV_4106 | - | 1e-60 | 75.17% (149) | Hsp20/alpha crystallin family protein |
| M. smegmatis MC2 155 | MSMEG_5611 | - | 5e-28 | 43.45% (145) | spore protein |
| M. thermoresistible (build 8) | TH_0596 | - | 2e-28 | 44.68% (141) | spore protein |
| M. ulcerans Agy99 | MUL_2232 | - | 3e-80 | 99.33% (149) | molecular chaperone (small heat shock protein) |
| M. vanbaalenii PYR-1 | Mvan_4943 | - | 2e-27 | 42.48% (153) | heat shock protein Hsp20 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_2991|M.marinum_M ----------MLMRTDPFRDLDRFAQQVL----GTSARPAVMPMD-----
MUL_2232|M.ulcerans_Agy99 ----------MLMRTDPFRDLDRFAQQVL----GTSARPAVMPMD-----
MLBr_01795|M.leprae_Br4923 ----------MLMRTDPFRELDRFAEQVL----GTSARPAVMPMD-----
MAV_4106|M.avium_104 ----------MLMRSDPFRELDRLTNQVL----GTATRPAVMPMD-----
MAB_3467c|M.abscessus_ATCC_199 ----------MLMRTDPFRDLDRFTQQVL----GTAARPALMPMD-----
Mflv_1805|M.gilvum_PYR-GCK -----------MLRFDPFSDIDALTRGLLTSQTGSNRTPRFMPMD-----
Mvan_4943|M.vanbaalenii_PYR-1 -----------MLRFDPFSDLDALTRGLLTSQTGSSRSPRFMPMD-----
MSMEG_5611|M.smegmatis_MC2_155 -----------MLRFDPFSDLDALTKGLLSTGTGSSRTPRFMPMD-----
TH_0596|M.thermoresistible__bu VAGLGSQEVIAVLRFDPFSDLDEWARSMLSTQVGSNRAPRFMPMD-----
Mb0257c|M.bovis_AF2122/97 -------MNNLALWSRPVWDVEPWDRWLRDFFGPAATTDWYRPVAGDFTP
Rv0251c|M.tuberculosis_H37Rv -------MNNLALWSRPVWDVEPWDRWLRDFFGPAATTDWYRPVAGDFTP
: *. ::: . : : *:
MMAR_2991|M.marinum_M ---AWREGDKFVVEFDLPGIDAD-SLDIDIERN-----VVTVRAERPA-V
MUL_2232|M.ulcerans_Agy99 ---AWREGDKFVVEFDLPGIDAD-SLDIDIERN-----VVTVRAERPA-V
MLBr_01795|M.leprae_Br4923 ---AWREGEEFVVEFDLPGIKAD-SLDIDIERN-----VVTVRAERPG-V
MAV_4106|M.avium_104 ---AWREGEHFVVEFDLPGIDAD-SLDIDIERN-----VLTVRAERPA-L
MAB_3467c|M.abscessus_ATCC_199 ---AWRDGNEFIVEFDLPGISGE-SLDLDIEHN-----VVTVRAERAP-V
Mflv_1805|M.gilvum_PYR-GCK ---LCKIDDHYVLTADLPGVDPG-SVDVDVDNG-----TLTISAHRTARS
Mvan_4943|M.vanbaalenii_PYR-1 ---LCKIDDHYVLTADLPGVDPG-SVDVNVDNG-----TLTISAHRTARS
MSMEG_5611|M.smegmatis_MC2_155 ---LCKIDDHYVLTADLPGVDPG-SVDVTVDNG-----MLTISAHRTARS
TH_0596|M.thermoresistible__bu ---LCKIDDHYVLTADLPGVDPG-SVDVSVENG-----MLTISAHRSARS
Mb0257c|M.bovis_AF2122/97 AAEIVKDGDDAVVRLELPGIDVDKDVNVELDPGQPVSRLVIRGEHRDEHT
Rv0251c|M.tuberculosis_H37Rv AAEIVKDGDDAVVRLELPGIDVDKDVNVELDPGQPVSRLVIRGEHRDEHT
: .:. :: :***:. .::: :: . : .*
MMAR_2991|M.marinum_M DPNRE---MLASERPRGVFSRQLVLGENLDTARIAASYTEGVLKLQIP--
MUL_2232|M.ulcerans_Agy99 DPNRE---MLASERPRGVFSRQLVLGENLDTARIAASYTEGVLKLQIP--
MLBr_01795|M.leprae_Br4923 DPDRE---MLAAERPRGVFNRQLVLGENLDTERILASYQEGVLKLSIP--
MAV_4106|M.avium_104 DPNRE---MLASERPRGVFSRELVLGDNLDTDKIEASYRDGVLSLHIP--
MAB_3467c|M.abscessus_ATCC_199 DPGRE---MLASERARGVFSRQLVLGDNLDTEKIAADYRNGVLRLRIP--
Mflv_1805|M.gilvum_PYR-GCK DESAQ---WLSSERFYGTYRRQLSLGDGIDTGAISATYENGVLTVTIP--
Mvan_4943|M.vanbaalenii_PYR-1 EESTQ---WLANERFFGSYRRQLSLGDGVDTAAISATYENGVLTVTIP--
MSMEG_5611|M.smegmatis_MC2_155 DESAQ---WLANERFFGSYRRQLSLGEGIDTSAIAATYENGVLTVTIP--
TH_0596|M.thermoresistible__bu DESAQ---WLANERFFGNFRRQISLGEGVDTSAIKATYENGVLTVTIP--
Mb0257c|M.bovis_AF2122/97 QDAGDKDGRTLREIRYGSFRRSFRLPAHVTSEAIAASYDAGVLTVRVAGA
Rv0251c|M.tuberculosis_H37Rv QDAGDKDGRTLREIRYGSFRRSFRLPAHVTSEAIAASYDAGVLTVRVAGA
: : * * : *.: * : : * * * *** : :.
MMAR_2991|M.marinum_M --VAEKAKPRKISITRG-AGDKTISENGAHPEVIEA--
MUL_2232|M.ulcerans_Agy99 --VAEKAKPRKISITRG-AGDKTISENVAHPEVIEA--
MLBr_01795|M.leprae_Br4923 --VAERAKPRKISVDRGNNGHQTIN-KTAH-EIIDA--
MAV_4106|M.avium_104 --VAEKAKPRKIAVGRG-EAPRAVT-ETAR-EVVNA--
MAB_3467c|M.abscessus_ATCC_199 --VAEKAKPRKIVVDRPADAVVIEGENDSARGKISA--
Mflv_1805|M.gilvum_PYR-GCK --VAAKARPRKIAINHT----KSQTSIEAN--TVDAE-
Mvan_4943|M.vanbaalenii_PYR-1 --VAEKAKPRKIEISHT----GNQKSIQPT--TVEAG-
MSMEG_5611|M.smegmatis_MC2_155 --MVERAKPRKIEIAHG----GEQKSITVEGETVSNDQ
TH_0596|M.thermoresistible__bu --LAESAKPRKIAVERA----GEQKTIEAA--TVDAE-
Mb0257c|M.bovis_AF2122/97 YKAPAETQAQRIAITK----------------------
Rv0251c|M.tuberculosis_H37Rv YKAPAETQAQRIAITK----------------------
::.::* : :