For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LAGTPNAVARMLSLLGDEWTLLIVQRALLGARRYGDFAAALPVSNTVLSGRLQSLTADGLLIRSQYQSNP PRSEYLPTSMGRSLWPMLTSIWEWERRWVPEHAEPLPRMHHATCGRPFQPVVTCRACDGPASGKDVAAQW GPSGSWERSIPSGTNRRRSSGRRSGAALLFPQTMSVVGDRWAFALLVAAFVGVGRFTDFQAQLGAPPATI ADRLSVFTDENILGQSGGRYGLTEKGLAFFPVLVCALAWAQRWLPSPEGPAVLLTHTACDRRFAPVLVCD QCGNRLRAEQVLPVS
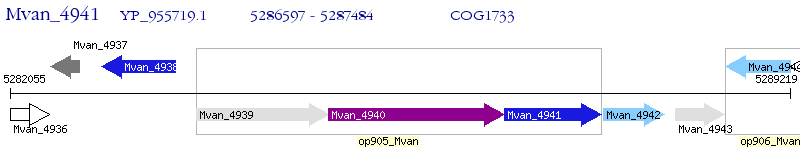
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_4941 | - | - | 100% (295) | HxlR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_2618 | - | 3e-10 | 26.89% (119) | HxlR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_1032 | - | 7e-08 | 32.00% (100) | HxlR family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3122 | - | 4e-21 | 35.29% (136) | putative transcriptional regulatory protein |
| M. gilvum PYR-GCK | Mflv_1806 | - | 1e-128 | 75.70% (284) | HxlR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv3095 | - | 4e-21 | 35.29% (136) | putative transcriptional regulatory protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3456 | - | 3e-15 | 31.40% (121) | HxlR family transcriptional regulator |
| M. marinum M | MMAR_1555 | - | 6e-18 | 33.58% (137) | transcriptional regulatory protein |
| M. avium 104 | MAV_4183 | - | 6e-10 | 29.17% (120) | putative transcriptional regulator family protein |
| M. smegmatis MC2 155 | MSMEG_5610 | - | 5e-94 | 57.24% (297) | putative transcriptional regulator family protein |
| M. thermoresistible (build 8) | TH_2405 | - | 9e-09 | 36.26% (91) | putative transcriptional regulator |
| M. ulcerans Agy99 | MUL_3475 | - | 4e-18 | 33.58% (137) | transcriptional regulatory protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3122|M.bovis_AF2122/97 --------MAVSDLSHRFEGESVGRALELVGERWTLLILREAFFGVRRFG
Rv3095|M.tuberculosis_H37Rv --------MAVSDLSHRFEGESVGRALELVGERWTLLILREAFFGVRRFG
MMAR_1555|M.marinum_M --------MGAADLSHRFEGESVGRALELVGERWTLLILREAFFGVQRFG
MUL_3475|M.ulcerans_Agy99 --------MGAADLSHRFEGESVGRALELVGERWTLLILREAFFGVQRFG
MAB_3456|M.abscessus_ATCC_1997 --------MDFTTMRN----CSISNALDVIGERWTLVALREIMLGNRRFE
Mvan_4941|M.vanbaalenii_PYR-1 ---------------MAGTPNAVARMLSLLGDEWTLLIVQRALLGARRYG
Mflv_1806|M.gilvum_PYR-GCK -------------------------MLGLLGDEWTLLIVQRALLGARRYG
MSMEG_5610|M.smegmatis_MC2_155 MDDPQPRTASGPTELAPGGTNAVGHMLALLGDEWSLLVMQQALLGATRYG
MAV_4183|M.avium_104 -MTLLQGPLADRDAWSAVGECAIEKTMAVVGTKSAMLIMREAYYGTTRFD
TH_2405|M.thermoresistible__bu -----------MASRTYGQHCALAKSLDLVGDRWTLLIVRELLDGPRRYS
: ::* . ::: ::. * *:
Mb3122|M.bovis_AF2122/97 QLARNLG-IPRPTLSSRLRMLVEVGLFDRVPYSS--DPERHEYRLTEAGR
Rv3095|M.tuberculosis_H37Rv QLARNLG-IPRPTLSSRLRMLVEVGLFDRVPYSS--DPERHEYRLTEAGR
MMAR_1555|M.marinum_M QLARNLN-IPRPTLSSRLRMLVNAGLLDRVPYSQ--DPERHEYRLTDTGH
MUL_3475|M.ulcerans_Agy99 QLARNLN-IPRPTLSSRLRMLVNAGLLDRVPYSQ--DPERHEYRLTDTGH
MAB_3456|M.abscessus_ATCC_1997 TIVRNTG-ASRDILATRLRKLVDAGVLEKRRYED--RPPRYEYLLTQSGQ
Mvan_4941|M.vanbaalenii_PYR-1 DFAAALP-VSNTVLSGRLQSLTADGLLIRSQYQS--NPPRSEYLPTSMGR
Mflv_1806|M.gilvum_PYR-GCK EFLDALP-VSNAVLTSRLQSLSATEMLSRNQYQS--NPPRFEYLVTPMSR
MSMEG_5610|M.smegmatis_MC2_155 QFMSRLP-ISNSVLTRRLGTLTADGLLRRHRYQD--RPPRDEYLVTARGR
MAV_4183|M.avium_104 DFARRVG-ITKAATSARLAELVELGLLKRRPYREPGQRSRDEYVLTQAGI
TH_2405|M.thermoresistible__bu DLLTALRPIATDMLATRLRHLEEHHLVRRRELPK--PASGTVYELTADGA
: . : ** * :. : . * * .
Mb3122|M.bovis_AF2122/97 DLFAAIVVLMQWGDEYLPRPEGPPIKLRHHTCGEHADPRLICTHCGEEIT
Rv3095|M.tuberculosis_H37Rv DLFAAIVVLMQWGDEYLPRPEGPPIKLRHHTCGEHADPRLICTHCGEEIT
MMAR_1555|M.marinum_M ELFAAIVVLMQWGDKHLPNPDGPPIVLRHH-CGQFVEPRLICAHCGEEIT
MUL_3475|M.ulcerans_Agy99 ELFAAIVVLMQWGDKHLPNPDGPPIVLRHH-CGQFVEPRLICAHCGEEIT
MAB_3456|M.abscessus_ATCC_1997 ALRPVLFALMEWGDKFVTK--GPPPSIWEHQCGSVLHIQPTCESCGEAVT
Mvan_4941|M.vanbaalenii_PYR-1 SLWPMLTSIWEWERRWVPEHAEPLPRMHHATCGRPFQPVVTCRACDGPAS
Mflv_1806|M.gilvum_PYR-GCK SLWPMLTSIWEWERRWVTDHAEPLPRMRHTGCGHEFAPRVVCRSCTAEAS
MSMEG_5610|M.smegmatis_MC2_155 ALWPILLSIWEWERRWVPEQRDALPGMRHISCGEEFHPLLSCSACEKEAN
MAV_4183|M.avium_104 DFMPVVWAMFEWGRRHLPG--RHRLQLTHLGCGAEVGVEIRCAEG-HLVP
TH_2405|M.thermoresistible__bu ALEAVVHAFIRWG-RSLLETRRAGDVVRPHWLARAVRAHVRPDRTGPPLS
: . : : .* . : : .
Mb3122|M.bovis_AF2122/97 ARNVTPEPGPGFKAKLASS-------------------------------
Rv3095|M.tuberculosis_H37Rv ARNVTPEPGPGFKAKLASS-------------------------------
MMAR_1555|M.marinum_M ARNVTPEAGPGFGDAVAAPN------------------------------
MUL_3475|M.ulcerans_Agy99 ARNVTPEAGPGYGDAVAAPN------------------------------
MAB_3456|M.abscessus_ATCC_1997 FDDLTPR-------RLGRVR------------------------------
Mvan_4941|M.vanbaalenii_PYR-1 GKDVAAQWGPSGSWERSIPSGTNRRRS-SGR----RSGAALLFPQTMSVV
Mflv_1806|M.gilvum_PYR-GCK GNDVVAQWGPSGSWQRSIPGGTQRRRS-SGH----RSGAAILFPQTMSVI
MSMEG_5610|M.smegmatis_MC2_155 PGEVVARWGPSGGWQRALPVATTRRRSGSGSGADDRDSPSGLFPQTMSVL
MAV_4183|M.avium_104 PDELGMRLAKKSG-------------------------------------
TH_2405|M.thermoresistible__bu VRLVTPEGAAAVRISADAVED---------------------LDDDTAVD
: .
Mb3122|M.bovis_AF2122/97 --------------------------------------------------
Rv3095|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1555|M.marinum_M --------------------------------------------------
MUL_3475|M.ulcerans_Agy99 --------------------------------------------------
MAB_3456|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_4941|M.vanbaalenii_PYR-1 GDRWAFALLVAAFVGVGRFTDFQAQLGAPPATIADRLSVFTDENILGQSG
Mflv_1806|M.gilvum_PYR-GCK GDRWAFAMLVAAFVGMRRFTDFQAQLGAPPTTVADRLTTFTDEGILVSGD
MSMEG_5610|M.smegmatis_MC2_155 GNRWACAILVAAFTGTTRFSTFQKLLAAPPGSVSDRLARFRANGIMTTSP
MAV_4183|M.avium_104 --------------------------------------------------
TH_2405|M.thermoresistible__bu VTLTGAAEVLAAALHPDRAAELVAQGRLRVDGRADALRRLAEVVTAPAGR
Mb3122|M.bovis_AF2122/97 --------------------------------------------------
Rv3095|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1555|M.marinum_M --------------------------------------------------
MUL_3475|M.ulcerans_Agy99 --------------------------------------------------
MAB_3456|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_4941|M.vanbaalenii_PYR-1 GR--YGLTEKGLAFFPVLVCALAWAQRWLPSPEGPAVLLTHTACDRRFAP
Mflv_1806|M.gilvum_PYR-GCK GG--YQFTDKGRAFFPVLVCALSWAQRWFPSPEGPAVILTHTACGRRFDP
MSMEG_5610|M.smegmatis_MC2_155 ERPEYLLTAKGEAFLPVLVTGLQWAQGWFSAPEGPAVNLTHTACDQVFHP
MAV_4183|M.avium_104 --------------------------------------------------
TH_2405|M.thermoresistible__bu AVPDRS--------------------------------------------
Mb3122|M.bovis_AF2122/97 --------------------
Rv3095|M.tuberculosis_H37Rv --------------------
MMAR_1555|M.marinum_M --------------------
MUL_3475|M.ulcerans_Agy99 --------------------
MAB_3456|M.abscessus_ATCC_1997 --------------------
Mvan_4941|M.vanbaalenii_PYR-1 VLVCDQCGNRLRAEQVLPVS
Mflv_1806|M.gilvum_PYR-GCK VLRCDQCQRHLRGAQIRPVP
MSMEG_5610|M.smegmatis_MC2_155 VLRCDRCKEPLTGNSVQTI-
MAV_4183|M.avium_104 --------------------
TH_2405|M.thermoresistible__bu --------------------