For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AKHVRLSRYSRYTGGPDPLAPPVDLREALEAIGQDVMEGTSPRRALSEMLRRGTQNMPGADKLAAEANRR RRELLQRNNLDGTLADIKKLLDEAVLAERKELARALDDDARFGELQLNSLSPSPAKAVQELSDYDWRSPE AREKYDQIKDLLGREMLDQRFAGMKEALENATDEDRQRVNDMLDDLNDLLDKHAKGQDSPQDFQDFMAKH GQFFPENPKNIDELLDSLAKRAAAAQRFRNSLSEQQRAELDALAQQAFGSPSLMNALDRLDAHLQSARPG EDWTGSQRFSGDDPLGMGEGAQALADIAELEQLAEQLSQSYSGATMDDVDLDMLARQLGDEAAVNARTLA ELERALMNQGFLDRGSDGQWRLSPKAMRQLGQAALRDVAQQLSGRHGERDTRRAGAAGELTGATRPWQFG DTEPWNVTRTVTNAVLRTAGTNAVLREAGTGETAGPIRITVDDVEISETETRTQAAVALLVDTSFSMVME NRWLPMKRTALALNHLVSTRFRSDALQIVAFGRYARTVTAAELTGLEGVYEQGTNLHHALALATRHLRRH PNAQPVVLVVTDGEPTAHLEDFDGDGRSAVFFDYPPHPRTIAHTVKGFDEVARMGAQVTIFRLGSDPGLA RFIDQVARRVGGRVVVPDLDGLGAAVVGDYLSAKRRR*
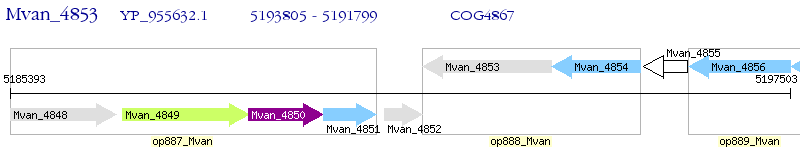
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_4853 | - | - | 100% (668) | von Willebrand factor, type A |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0984 | - | 0.0 | 83.92% (659) | hypothetical protein Mb0984 |
| M. gilvum PYR-GCK | Mflv_1878 | - | 0.0 | 92.31% (663) | von Willebrand factor, type A |
| M. tuberculosis H37Rv | Rv0959 | - | 0.0 | 83.92% (659) | hypothetical protein Rv0959 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_4540 | - | 0.0 | 86.10% (662) | hypothetical protein MMAR_4540 |
| M. avium 104 | MAV_1084 | - | 0.0 | 86.67% (645) | von Willebrand factor, type A |
| M. smegmatis MC2 155 | MSMEG_5511 | - | 0.0 | 88.59% (666) | von Willebrand factor, type A |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_4710 | - | 0.0 | 86.40% (662) | hypothetical protein MUL_4710 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4540|M.marinum_M ---------------MADRAKGHSTRYSAYTGGPDPLAPPVDLREALEQI
MUL_4710|M.ulcerans_Agy99 ---------------MADRAKGHSTRYSAYTGGPDPLAPPVDLREALEQI
MAV_1084|M.avium_104 ----------------------------------------MDLREALEQI
Mb0984|M.bovis_AF2122/97 MAKSDGDDPLRPASPRLRSSRRHSLRYSAYTGGPDPLAPPVDLRDALEQI
Rv0959|M.tuberculosis_H37Rv MAKSDGDDPLRPASPRLRSSRRHSLRYSAYTGGPDPLAPPVDLRDALEQI
Mvan_4853|M.vanbaalenii_PYR-1 -----------MAK------HVRLSRYSRYTGGPDPLAPPVDLREALEAI
Mflv_1878|M.gilvum_PYR-GCK --------------------MSRTSRYSRYTGGPDPLAPPVDLRDALEAI
MSMEG_5511|M.smegmatis_MC2_155 -----------MADNASGRGHGHTSRYSRYTGGPDPLAPPIDLREALEQI
:***:*** *
MMAR_4540|M.marinum_M GQDVMAGTSPRRALSELLRRGTKSMPGADRLASEANRRRRELLRRNNLDG
MUL_4710|M.ulcerans_Agy99 GQDVMAGTSPRRALSELLRRGTKSMPGADRLASEANRRRRELLRRNNLDG
MAV_1084|M.avium_104 GQDVMAGTSPRRALSELLRRGTKNMPGADRLAAEANRRRRELLRRNNLDG
Mb0984|M.bovis_AF2122/97 GQDVMAGASPRRALSELLRRGTRNLTGADRLAAEVNRRRRELLRRNNLDG
Rv0959|M.tuberculosis_H37Rv GQDVMAGASPRRALSELLRRGTRNLTGADRLAAEVNRRRRELLRRNNLDG
Mvan_4853|M.vanbaalenii_PYR-1 GQDVMEGTSPRRALSEMLRRGTQNMPGADKLAAEANRRRRELLQRNNLDG
Mflv_1878|M.gilvum_PYR-GCK GQDVMEGTSPRRALSELLRRGTKNMPGADKLAAEANRRRRELLQRNNLDG
MSMEG_5511|M.smegmatis_MC2_155 GQNVMEGSSPRRALSELMRRGTQNMRGADRLAAEANRRRRELLQRNNLDG
**:** *:********::****:.: ***:**:*.********:******
MMAR_4540|M.marinum_M TLQEIKKLLDEAVLAERKELARALDDDARFSELQLEALPPSPAKAVQELS
MUL_4710|M.ulcerans_Agy99 TLQEIKKLLDEAVLAERKELARALDDDARFSELQLEALPPSPAKAVQELS
MAV_1084|M.avium_104 TLQEIKKLLDEAVLAERKELARALDDDARFGELQLDALPASPAKAVQELS
Mb0984|M.bovis_AF2122/97 TLQEIKKLLDEAVLAERKELARALDDDARFAELQLDALPASPAKAVQELA
Rv0959|M.tuberculosis_H37Rv TLQEIKKLLDEAVLAERKELARALDDDARFAELQLDALPASPAKAVQELA
Mvan_4853|M.vanbaalenii_PYR-1 TLADIKKLLDEAVLAERKELARALDDDARFGELQLNSLSPSPAKAVQELS
Mflv_1878|M.gilvum_PYR-GCK TLTEIKKLLDEAVLAERKELARALDDDARFGELQLESLSPSPAKAVQELA
MSMEG_5511|M.smegmatis_MC2_155 TLQEIKKLLDEAVLAERKELARALDDDARFGELQLDALSPSPAKAVQELA
** :**************************.****::*..*********:
MMAR_4540|M.marinum_M EYDWRSGEAREKYDQIKDLLGRELLDQRFAGMKQALEGATDEDRERVNEM
MUL_4710|M.ulcerans_Agy99 EYDWRSGEAREKYDQIKDLLGRELLDQRFAGMKQALEGATDEDRQRVNEM
MAV_1084|M.avium_104 EYNWRSSEAREKYDQIKDLLGREMLDQRFAGMKQALQGATDEDRQRVSDM
Mb0984|M.bovis_AF2122/97 EYRWRSGQAREKYEQIKDLLGRELLDQRFAGMKQALAGATDDDRRRVTEM
Rv0959|M.tuberculosis_H37Rv EYRWRSGQAREKYEQIKDLLGRELLDQRFAGMKQALAGATDDDRRRVTEM
Mvan_4853|M.vanbaalenii_PYR-1 DYDWRSPEAREKYDQIKDLLGREMLDQRFAGMKEALENATDEDRQRVNDM
Mflv_1878|M.gilvum_PYR-GCK EYDWRSAEAREKYDQIKDLMGREMLDQRFAGMKEALENATDEDRQRVNDM
MSMEG_5511|M.smegmatis_MC2_155 EYDWRSQEAREKYEQIKDLMGREMLDQRFAGMKQALENATDEDRQRVNDM
:* *** :*****:*****:***:*********:** .***:**.**.:*
MMAR_4540|M.marinum_M LDDLNNLLDKHARGQDTQQDFQDFMDKHGEFFPENPSNVEELLDSLAKRA
MUL_4710|M.ulcerans_Agy99 LDDLNNLLDKHARGQDTQQDFQDFMDKHGEFFPENPSNVEELLDSLAKRA
MAV_1084|M.avium_104 LNDLNDLLDKHAKGQDSQQDFDDFMAKHGEFFPESPRNVEELLDSLAKRA
Mb0984|M.bovis_AF2122/97 LDDLNDLLDKHARGEDTQRDFDEFMTKHGEFFPENPRNVEELLDSLAKRA
Rv0959|M.tuberculosis_H37Rv LDDLNDLLDKHARGEDTQRDFDEFMTKHGEFFPENPRNVEELLDSLAKRA
Mvan_4853|M.vanbaalenii_PYR-1 LDDLNDLLDKHAKGQDSPQDFQDFMAKHGQFFPENPKNIDELLDSLAKRA
Mflv_1878|M.gilvum_PYR-GCK LDDLNELLDKHARGEDTDQGFRDFMAKHGEFFPENPQNIDELLDSLAQRA
MSMEG_5511|M.smegmatis_MC2_155 LDDLNNLLDKHAAGQDTQQDFQDFMDKHGEFFPENPQNIDELLDSLAKRA
*:***:****** *:*: :.* :** ***:****.* *::*******:**
MMAR_4540|M.marinum_M AAAQRFRNSLSQEQRDELDALAQQAFGSPSLMQALNRLDAHLQEARPGED
MUL_4710|M.ulcerans_Agy99 AAAQRFRNSLSQEQRDELDALAQQAFGSPSLMQALNRLDAHLQEARPGED
MAV_1084|M.avium_104 AAAQRFRNSLSAEQRAELDALAQQAFGSPDLMQALNRLDAHLQAARPGED
Mb0984|M.bovis_AF2122/97 AAAQRFRNSLSQEQRDELDALAQQAFGSPALMRALDRLDAHLQAARPGED
Rv0959|M.tuberculosis_H37Rv AAAQRFRNSLSQEQRDELDALAQQAFGSPALMRALDRLDAHLQAARPGED
Mvan_4853|M.vanbaalenii_PYR-1 AAAQRFRNSLSEQQRAELDALAQQAFGSPSLMNALDRLDAHLQSARPGED
Mflv_1878|M.gilvum_PYR-GCK AAAQRFRNSLTEQQRAELDSLAQQAFGSPSLMNALNRLDSHLQAARPGED
MSMEG_5511|M.smegmatis_MC2_155 AAAQRFRNSLSPDQRAELDALAQQAFGSPSLMNALNRLDAHLQSARPGED
**********: :** ***:********* **.**:***:*** ******
MMAR_4540|M.marinum_M WTGQQQFSGDNPFGMGEGTQALADIAELEQLAEQLSQAYPGATMDDVDLD
MUL_4710|M.ulcerans_Agy99 WTGQQRFSGDNPFGMGEGTQALADIAELEQLAEQLSQAYPGATMDDVDLD
MAV_1084|M.avium_104 WGGSEQFSGDNPFGMGEGTQALADIAELEQLAEQLSQSYPGATMDDVDLD
Mb0984|M.bovis_AF2122/97 WTGSQQFSGDNPFGMGEGTQALADIAELEQLAEQLSQSYPGASMDDVDLD
Rv0959|M.tuberculosis_H37Rv WTGSQQFSGDNPFGMGEGTQALADIAELEQLAEQLSQSYPGASMDDVDLD
Mvan_4853|M.vanbaalenii_PYR-1 WTGSQRFSGDDPLGMGEGAQALADIAELEQLAEQLSQSYSGATMDDVDLD
Mflv_1878|M.gilvum_PYR-GCK WSGSQRFSGDDPLGMGEGAQALADIAELEQLAEQLSQSYSGASMDDVDLD
MSMEG_5511|M.smegmatis_MC2_155 WNGAQRFSGDDPLGMGEGAQALADIAELEQLADALSQSYAGATMDDVDLE
* * ::****:*:*****:*************: ***:*.**:******:
MMAR_4540|M.marinum_M ALARQLGDQAAIDARTLAELERALVNQGFFDRGSDGQWRLSPKAMRRLGE
MUL_4710|M.ulcerans_Agy99 ALARQLGDQAAIDARTLAELERALVNQGFFDRGSDGQWRLSPKAMRRLGE
MAV_1084|M.avium_104 ALARQLGDQAAVDARTLAELERALVNQGFLDRGSDGQWRLSPKAMRRLGE
Mb0984|M.bovis_AF2122/97 ALARQLGDQAAVDARTLAELERALVNQGFLDRGSDGQWRLSPKAMRRLGE
Rv0959|M.tuberculosis_H37Rv ALARQLGDQAAVDARTLAELERALVNQGFLDRGSDGQWRLSPKAMRRLGE
Mvan_4853|M.vanbaalenii_PYR-1 MLARQLGDEAAVNARTLAELERALMNQGFLDRGSDGQWRLSPKAMRQLGQ
Mflv_1878|M.gilvum_PYR-GCK MLARQLGDEAAVDARTLSELERALMNQGFLDRGSDGQWRLSPKAMRQLGQ
MSMEG_5511|M.smegmatis_MC2_155 ALARQLGDEAAVDARTLAELERALVNQGFLDRGSDGQWRLSPKAMRQLGQ
*******:**::****:******:****:****************:**:
MMAR_4540|M.marinum_M TALRDVVQQLSGRRGERDHRRAGAAGELTGATRPWQFGDTEPWHISRTLT
MUL_4710|M.ulcerans_Agy99 TALRDVVQQLSGRRGERDHRRAGAAGELTGATRPWQFGDTEPWHISRTLT
MAV_1084|M.avium_104 TALRDVAQQLSGRRGERDHRRAGAAGELTGATRPWQFGDTEPWNISRTLT
Mb0984|M.bovis_AF2122/97 TALRDVAQQLSGRHGERDHRRAGAAGELTGATRPWQFGDTEPWHVARTLT
Rv0959|M.tuberculosis_H37Rv TALRDVAQQLSGRHGERDHRRAGAAGELTGATRPWQFGDTEPWHVARTLT
Mvan_4853|M.vanbaalenii_PYR-1 AALRDVAQQLSGRHGERDTRRAGAAGELTGATRPWQFGDTEPWNVTRTVT
Mflv_1878|M.gilvum_PYR-GCK AALRDVAQQLSGRHGERDTRRAGAAGELTGATRPWQFGDTEPWNVTRTLT
MSMEG_5511|M.smegmatis_MC2_155 TALRDVAQQLSGRHGERDTRRAGAAGELTGATRPWQFGDTEPWNVTRTVT
:*****.******:**** ************************:::**:*
MMAR_4540|M.marinum_M NAVLRQAGTAPS------GGGAGPIRITVDDVEVSETETRTQAAVALLVD
MUL_4710|M.ulcerans_Agy99 NAVLRQAGTAPP------GGAAGPIRITVDDVEVSETETRTQAAVALLVD
MAV_1084|M.avium_104 NAVLRQAGTATL------DGPDGRLKITVDDVEVSETETRTQAAVALLVD
Mb0984|M.bovis_AF2122/97 NAVLRQAAAVH-----------DRIRITVEDVEVAETETRTQAAVALLVD
Rv0959|M.tuberculosis_H37Rv NAVLRQAAAVH-----------DRIRITVEDVEVAETETRTQAAVALLVD
Mvan_4853|M.vanbaalenii_PYR-1 NAVLRTAGTNAVLREAGTGETAGPIRITVDDVEISETETRTQAAVALLVD
Mflv_1878|M.gilvum_PYR-GCK NAVLREAGT-------AKDRPGGRLRITVDDVEISETETRTQAAVALLVD
MSMEG_5511|M.smegmatis_MC2_155 NAVLRQAGT---------GLVERPIRFAVDDVEISETETRTQACVALLVD
***** *.: ::::*:***::********.******
MMAR_4540|M.marinum_M TSFSMVMENRWLPMKRTALALNHLVCTRFRSDALQIIAFGRYARTVTAAE
MUL_4710|M.ulcerans_Agy99 TSFSMVMENRWLPMKRTALALNHLVCTRFRSDALQIIAFGRYARTVTAAE
MAV_1084|M.avium_104 TSFSMVMENRWLPMKQTALALNHLVCTRFRSDALQIIAFGRYARTVTAAE
Mb0984|M.bovis_AF2122/97 TSFSMVMENRWLPMKRTALALHHLVCTRFRSDALQIIAFGRYARTVTAAE
Rv0959|M.tuberculosis_H37Rv TSFSMVMENRWLPMKRTALALHHLVCTRFRSDALQIIAFGRYARTVTAAE
Mvan_4853|M.vanbaalenii_PYR-1 TSFSMVMENRWLPMKRTALALNHLVSTRFRSDALQIVAFGRYARTVTAAE
Mflv_1878|M.gilvum_PYR-GCK TSFSMVMENRWLPMKRTALALNHLVSTRFRSDALQIVAFGRYARTVTAAE
MSMEG_5511|M.smegmatis_MC2_155 TSFSMVMENRWLPMKRTALALNHLVSTRFRSDALQIIAFGRYARTVSAAE
***************:*****:***.**********:*********:***
MMAR_4540|M.marinum_M LTGLEGVYEQGTNLHHALALAGRHLRRHPNAQPVVLVVTDGEPTAHLEDF
MUL_4710|M.ulcerans_Agy99 LTGLEGVYEQGTNLHHALALAGRHLRRHPNAQPVVLVVTDGEPTAHLEDF
MAV_1084|M.avium_104 LTGLEGVYEQGTNLHHALALAGRHLRRHPNAQPVVLVVTDGEPTAHLEDF
Mb0984|M.bovis_AF2122/97 LTGLAGVYEQGTNLHHALALAGRHLRRHAGAQPVVLVVTDGEPTAHLEDF
Rv0959|M.tuberculosis_H37Rv LTGLAGVYEQGTNLHHALALAGRHLRRHAGAQPVVLVVTDGEPTAHLEDF
Mvan_4853|M.vanbaalenii_PYR-1 LTGLEGVYEQGTNLHHALALATRHLRRHPNAQPVVLVVTDGEPTAHLEDF
Mflv_1878|M.gilvum_PYR-GCK LTGLEGVYEQGTNLHHALALATRHLRRHPNAQPVILVVTDGEPTAHLEDF
MSMEG_5511|M.smegmatis_MC2_155 LTALEGVYEQGTNLHHALALASRHLRRHPNAQPVVLVVTDGEPTAHLENF
**.* **************** ******..****:*************:*
MMAR_4540|M.marinum_M -AGEGAGSTVFFDYPPHPRTIAHTVRGFDDMARLGAQITIFRLGSDRGLA
MUL_4710|M.ulcerans_Agy99 -SGEGAGSAVFFDYPPHPRTIAHTVRGFDDMARLDAQITIFRLGSDRGLA
MAV_1084|M.avium_104 -EGE--GTTVFFDYPPHPRTIAHTVRGFDEMARLGAQITIFRLGSDPGLA
Mb0984|M.bovis_AF2122/97 -DGD--GTSVFFDYPPHPRTIAHTVRGFDDMARLGAQVTIFRLGSDPGLA
Rv0959|M.tuberculosis_H37Rv -DGD--GTSVFFDYPPHPRTIAHTVRGFDDMARLGAQVTIFRLGSDPGLA
Mvan_4853|M.vanbaalenii_PYR-1 --DGDGRSAVFFDYPPHPRTIAHTVKGFDEVARMGAQVTIFRLGSDPGLA
Mflv_1878|M.gilvum_PYR-GCK --DGDGRSSVFFDYPPHPRTIAHTVRGFDEVARLGAQVTIFRLGSDPGLA
MSMEG_5511|M.smegmatis_MC2_155 RISEDEGAQVFFDYPPHPRTIAHTVRGFDEVARLGAQVTIFRLGSDPGLA
. : ****************:***::**:.**:******** ***
MMAR_4540|M.marinum_M RFIDQVARRVEGRVVEPDLDGLGAAVVGDYLRSRRRR
MUL_4710|M.ulcerans_Agy99 RFIDQVARRVEGRVVEPDLDGLGAAVVGDYLRSRRRR
MAV_1084|M.avium_104 RFIDQVARRVEGRVVVPDLDGLGAAVVGDYLRSRRRR
Mb0984|M.bovis_AF2122/97 RFIDQVARRVQGRVVVPDLDGLGAAVVGDYLRFRRR-
Rv0959|M.tuberculosis_H37Rv RFIDQVARRVQGRVVVPDLDGLGAAVVGDYLRFRRR-
Mvan_4853|M.vanbaalenii_PYR-1 RFIDQVARRVGGRVVVPDLDGLGAAVVGDYLSAKRRR
Mflv_1878|M.gilvum_PYR-GCK RFIDQVARRVGGRVVVPDLDGLGAAVVGDYLSAKRRR
MSMEG_5511|M.smegmatis_MC2_155 RFIDQVARRVQGRVVVPDLDGLGAAVVGDYLRSRRR-
********** **** *************** :**