For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AADNAGRWPGQPPGPPAPTHPASSVSLRWRVMLLAMSMVVISVVLMAVAVFAVTSRALYDDIDNQLRSRA QMLIESRSLDIDPGKAIEGTAYSDMNAMFYIPGRSKYTANQQGQTLPVGQPEQDVMDGTLLLSLRTVEHQ RVLAIRLASGNTLLLSKSLAPTGKVLKRLGTVLLIVGGLGVAVAAIAGGMVASAGLRPVGRLTQAAERVA RTDDLRPIPVIGNDELARLTETFNMMLRALAESRERQARLVTDAGHELRTPLTSLRTNVELLMESMKPGA PRIPEEDMAELRTDVIAQIEEMSTLVGDLVDLTRDDAGNAVHETVEITEVIDRSLERVRRRRNDIQFDVA VTPWQVYGDAAGLGRAVLNLLDNAAKWSPPGGRVGVGLTQIDALHAELVVSDRGPGIPPQERALVFERFF RSTSARSMPGSGLGLAIVKQVVLKHGGTLRIEDTVPGGTPPGTAMHVVLPGRPSPAGSDEAER*
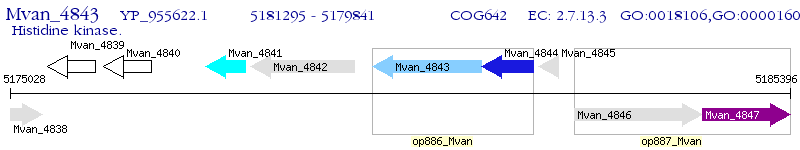
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_4843 | - | - | 100% (484) | integral membrane sensor signal transduction histidine kinase |
| M. vanbaalenii PYR-1 | Mvan_4428 | - | 1e-32 | 35.13% (353) | integral membrane sensor signal transduction histidine kinase |
| M. vanbaalenii PYR-1 | Mvan_5010 | - | 6e-32 | 29.31% (447) | integral membrane sensor signal transduction histidine kinase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1008 | mprB | 0.0 | 75.16% (459) | two component sensor kinase MprB |
| M. gilvum PYR-GCK | Mflv_1888 | - | 0.0 | 85.08% (476) | integral membrane sensor signal transduction histidine kinase |
| M. tuberculosis H37Rv | Rv0982 | mprB | 0.0 | 75.16% (459) | two component sensor kinase MprB |
| M. leprae Br4923 | MLBr_00175 | - | 0.0 | 73.16% (462) | putative two-component system sensor kinase |
| M. abscessus ATCC 19977 | MAB_1077 | - | 1e-179 | 70.87% (460) | two component sensor kinase MprB |
| M. marinum M | MMAR_4528 | mprB | 0.0 | 77.27% (462) | two-component sensor kinase MprB |
| M. avium 104 | MAV_1095 | - | 0.0 | 75.22% (460) | sensor histidine kinase |
| M. smegmatis MC2 155 | MSMEG_5487 | - | 0.0 | 76.63% (475) | sensor histidine kinase |
| M. thermoresistible (build 8) | TH_2428 | - | 0.0 | 78.97% (466) | PUTATIVE integral membrane sensor signal transduction histidine |
| M. ulcerans Agy99 | MUL_4700 | mprB | 0.0 | 77.27% (462) | two component sensor kinase MprB |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_4843|M.vanbaalenii_PYR-1 MAAD--------NAGRWPGQPPGPPAPTHPASS-VSLRWRVMLLAMSMVV
Mflv_1888|M.gilvum_PYR-GCK MAGDP-------NTATWPPQPPGPPPP--PASS-ISLRWRVMLLAMSMAV
MSMEG_5487|M.smegmatis_MC2_155 MASATCCGKRLRSEMAFPPNS-WRPTGPLPTSS-LSLRWRVMMLAMSMVA
TH_2428|M.thermoresistible__bu ----------MTFAQPYPPSPPQRQATSQPASS-LSLRWRVMLLAMSMVA
MLBr_00175|M.leprae_Br4923 ----------------MVRFAWRRRASLRATSS-LSLRWRVMLLAMSMVA
MAV_1095|M.avium_104 ----------------------------------------MMLLAMSMVA
Mb1008|M.bovis_AF2122/97 ----------------MWWFRRRDRAPLRATSS-LSLRWRVMLLAMSMVA
Rv0982|M.tuberculosis_H37Rv ----------------MWWFRRRDRAPLRATSS-LSLRWRVMLLAMSMVA
MMAR_4528|M.marinum_M ----------------MVGFRRGPRAPLRATSS-LSLRWRVMLLAMSMVA
MUL_4700|M.ulcerans_Agy99 ----------------MVGFRRGPRAPLRATSS-LSLRWRVMLLAMSMVA
MAB_1077|M.abscessus_ATCC_1997 -------MSTLRMPRRMRDSSTAIFTPLRATSSSVSLRWRVMLLAMSMVA
:*:*****..
Mvan_4843|M.vanbaalenii_PYR-1 ISVVLMAVAVFAVTSRALYDDIDNQLRSRAQMLIESRSLDIDPGKAIEGT
Mflv_1888|M.gilvum_PYR-GCK IPVVLMGVAGYAVVSRALYDDIDDQLRTRAQMLIESGSLDADPGKAIEGT
MSMEG_5487|M.smegmatis_MC2_155 LVVVLMAVAVYAVVSRALYDDLDNQLHSRARLLIESGSLAADPGKAIEGT
TH_2428|M.thermoresistible__bu MVVVLMSIALYAVVSRALYNDIDNQLHSRARLLIESGSLAADPGRAIEGT
MLBr_00175|M.leprae_Br4923 MVVVLMAFAVYVVISAALYSDIDNQLQSRAQLLIASGSLAADPGKAIEGT
MAV_1095|M.avium_104 MVVVLMAFAVYAVISAALYSDIDNQLQSRAQLLIASGSLAADPGKAIEGT
Mb1008|M.bovis_AF2122/97 MVVVLMSFAVYAVISAALYSDIDNQLQSRAQLLIASGSLAADPGKAIEGT
Rv0982|M.tuberculosis_H37Rv MVVVLMSFAVYAVISAALYSDIDNQLQSRAQLLIASGSLAADPGKAIEGT
MMAR_4528|M.marinum_M MVVVLMSFAVYAVISAALYSDIDNQLQSRAQLLIASGSLAADPGKAIEGT
MUL_4700|M.ulcerans_Agy99 MVVVLMSFAVYAVISAALYSDIDNQLQSRAQLLIASGSLAADPGKAIEGT
MAB_1077|M.abscessus_ATCC_1997 MVVVLMAVASYLLVSTALYRDVDKQLNDRANLLIGSGLLSVDPRRAIEGT
: ****..* : : * *** *:*.**. **.:** * * ** :*****
Mvan_4843|M.vanbaalenii_PYR-1 AYS-DMNAMFYIPGRSKYTANQQGQTLPVGQPEQDVMDGTLLLSLRTVEH
Mflv_1888|M.gilvum_PYR-GCK AYS-DMNAMFYIPGRSKYTAKQQGQTLPVGRPEQDVMDGTLWMSLRTVEH
MSMEG_5487|M.smegmatis_MC2_155 AYS-DVNAMLVIPGRSIYTANQQGQTLPLGEPEKDVISGELLMSLRTANH
TH_2428|M.thermoresistible__bu AYS-DVNAMLVNPGRSIYTANQQGEALPLGEPERQVLSGELVMSLRTADQ
MLBr_00175|M.leprae_Br4923 AYS-DVNAMLVNPGHSIYTANQPGQTLPVGTAEKAVIRGELFMSQRTASD
MAV_1095|M.avium_104 AYS-DVNAMLVNPGHAIYTAQQPGQTLPVGSPEKAVIHGELFMSRRTAGD
Mb1008|M.bovis_AF2122/97 AYS-DVNAMLVNPGQSIYTAQQPGQTLPVGAAEKAVIRGELFMSRRTTAD
Rv0982|M.tuberculosis_H37Rv AYS-DVNAMLVNPGQSIYTAQQPGQTLPVGAAEKAVIRGELFMSRRTTAD
MMAR_4528|M.marinum_M AYS-DVNAMLVNPGRSIYTANQPGQTLPVGAPEKAVIHGDLFLSRRTVSD
MUL_4700|M.ulcerans_Agy99 AYS-DVNAMLVNPGRSIYTANQPGQTLPVGAPEKAVIHGDLFLSRRTVSD
MAB_1077|M.abscessus_ATCC_1997 AYSADVNAKLIFPGLITFSATQQGQTIPYGTPEQAVTRGDLQQSLRTVNH
*** *:** : ** ::* * *:::* * .*: * * * * **. .
Mvan_4843|M.vanbaalenii_PYR-1 QRVLAIRLASGNTLLLSKSLAPTGKVLKRLGTVLLIVGGLGVAVAAIAGG
Mflv_1888|M.gilvum_PYR-GCK QRVLAVHLASGNSLLLSKSLAPTGQVLKRLGTVLLIVGGIGVVVAAIAGG
MSMEG_5487|M.smegmatis_MC2_155 QRVLAVHLANGSSLLISKSLAPTVQVLRRLGTVLLIVGGIGVAVAAIAGG
TH_2428|M.thermoresistible__bu QRVLAIHLSNGSSLLISKSLAPTMEVLRRLGTVLLIVGGIGVAVAAIAGG
MLBr_00175|M.leprae_Br4923 QRILAIHLPNDSSLLISKSLRPTEAVMTKLRWVLLIVGSLGVAVAAVAGG
MAV_1095|M.avium_104 QRILAVHLQNGTSLLISKSLKPTEAVMNKLRWVLLIVGGVGVAVAAVAGG
Mb1008|M.bovis_AF2122/97 QRVLAIRLTNGSSLLISKSLKPTEAVMNKLRWVLLIVGGIGVAVAAVAGG
Rv0982|M.tuberculosis_H37Rv QRVLAIRLTNGSSLLISKSLKPTEAVMNKLRWVLLIVGGIGVAVAAVAGG
MMAR_4528|M.marinum_M QRVLAIHLPNGSSLLISKSLKPTEAVMTKLRAVLLIVGGVGVAVAAVAGG
MUL_4700|M.ulcerans_Agy99 QRVLAIHLPNGSSLLISKSLKPTEAVMTKLRAVLLIVGGVGVAVAAVAGG
MAB_1077|M.abscessus_ATCC_1997 QRVLALRLDNGSTLIMAKSLDPTDTVLRRLGWVLLIVGGVGVAVAAAAGA
**:**::* ...:*:::*** ** *: :* ******.:**.*** **.
Mvan_4843|M.vanbaalenii_PYR-1 MVASAGLRPVGRLTQAAERVARTDDLRPIPVIGNDELARLTETFNMMLRA
Mflv_1888|M.gilvum_PYR-GCK MVARAGLRPVARLTQAAERVARTDDLHPIPVVGNDELARLTETFNMMLRA
MSMEG_5487|M.smegmatis_MC2_155 AVARAGLRPVGRLTEAAERVARTDDLRPIPVVGSDELARLTEAFNMMLRA
TH_2428|M.thermoresistible__bu MVARAGLRPVSRLTEAAERVARTDDLRPIPVVGSDELARLTEAFNMMLRA
MLBr_00175|M.leprae_Br4923 MVTRAGLRPVGRLTEAAERVARTDDLRPIPVFGSDELARLTEAFNLMLRA
MAV_1095|M.avium_104 MVTRAGLRPVARLTEAAERVARTDDLRPIPVFGSDELARLTESFNLMLRA
Mb1008|M.bovis_AF2122/97 MVTRAGLRPVGRLTEAAERVARTDDLRPIPVFGSDELARLTEAFNLMLRA
Rv0982|M.tuberculosis_H37Rv MVTRAGLRPVGRLTEAAERVARTDDLRPIPVFGSDELARLTEAFNLMLRA
MMAR_4528|M.marinum_M MVTRAGLRPVGRLTEAAERVARTDDLRPIPVFGSDELARLTEAFNLMLRA
MUL_4700|M.ulcerans_Agy99 MVTRAGLRPVGRLTEAAERVARTDDLRPIPVFGSDELARLTEAFNLMLRA
MAB_1077|M.abscessus_ATCC_1997 MVAGTGLRPVARLTDAAERVARTDDLRPIAVTGSDELARLTISFNTMLRA
*: :*****.***:***********:**.* *.******* :** ****
Mvan_4843|M.vanbaalenii_PYR-1 LAESRERQARLVTDAGHELRTPLTSLRTNVELLMESMKPGAPRIPEEDMA
Mflv_1888|M.gilvum_PYR-GCK LTESRDRQSRLVADAGHELRTPLTSLRTNVELLMSSMKPGAPRIPEEDMD
MSMEG_5487|M.smegmatis_MC2_155 LAESRERQARLVSDAGHELRTPLTSLRTNVELLMAAQEPGAPPLPEDEMA
TH_2428|M.thermoresistible__bu LAESRERQARLVADAGHELRTPLTSLRTNVELLMASQQPGAPGLPEEEMA
MLBr_00175|M.leprae_Br4923 LAESRERQARLVTDAGHELRTPLTSLRTNVELLIASMAPEAPRLPDQEMA
MAV_1095|M.avium_104 LAESRERQARLVTDAGHELRTPLTSLRTNVELLMASMEPGAPRLPEQEMV
Mb1008|M.bovis_AF2122/97 LAESRERQARLVTDAGHELRTPLTSLRTNVELLMASMAPGAPRLPKQEMV
Rv0982|M.tuberculosis_H37Rv LAESRERQARLVTDAGHELRTPLTSLRTNVELLMASMAPGAPRLPKQEMV
MMAR_4528|M.marinum_M LAESRERQARLVSDAGHELRTPLTSLRTNVELLMASMEPGAPRLPEQEMV
MUL_4700|M.ulcerans_Agy99 LAESRERQARLVSDAGHELRTPLTSLRTNVELLMASMEPGAPRLPEQEMV
MAB_1077|M.abscessus_ATCC_1997 LAESRDRQARLVTDAGHELRTPLTSLRTNTELLMAAMQPGAPRLPDEEMQ
*:***:**:***:****************.***: : * ** :*.::*
Mvan_4843|M.vanbaalenii_PYR-1 ELRTDVIAQIEEMSTLVGDLVDLTRDDAGNAVHETVEITEVIDRSLERVR
Mflv_1888|M.gilvum_PYR-GCK DLRSDVIGQIEELSTLVGDLVDLTRDEAGNAVHDTVEMSEVIERSLERVR
MSMEG_5487|M.smegmatis_MC2_155 GLRADVIAQIEELSTLVGDLVDLTREDAGGITPEPVDMADVIDRSLERVR
TH_2428|M.thermoresistible__bu GLRADVIAQIEELSTLVGDLVDLTRDDAGGVIHEPVDIADVVDRSLERVR
MLBr_00175|M.leprae_Br4923 DLRADVLAQIEELSTLVGDLVDLTRDDAGQVVHEPIDMSEVLYRSLERVR
MAV_1095|M.avium_104 ELRADVLAQIEELSTLVGDLVDLTRDDAGQVVHEPVDMSDVIDRSLERVR
Mb1008|M.bovis_AF2122/97 DLRADVLAQIEELSTLVGDLVDLSRGDAGEVVHEPVDMADVVDRSLERVR
Rv0982|M.tuberculosis_H37Rv DLRADVLAQIEELSTLVGDLVDLSRGDAGEVVHEPVDMADVVDRSLERVR
MMAR_4528|M.marinum_M GLREDVVAQIEELSTLVGDLVDLTRGDAGVVVHEPVDMAEVVDRSLERVR
MUL_4700|M.ulcerans_Agy99 GLREDVVAQIEELSTLVGDLVDLTRGDAGVVVHEPVDMAEVVDRSLERVR
MAB_1077|M.abscessus_ATCC_1997 ALQQDVIAQIEELSTLVGDLVDLARGDGADIVHEPVDMVEVVEGTLERVR
*: **:.****:**********:* :.. :.::: :*: :*****
Mvan_4843|M.vanbaalenii_PYR-1 RRRNDIQFDVAVTPWQVYGDAAGLGRAVLNLLDNAAKWSPPGGRVGVGLT
Mflv_1888|M.gilvum_PYR-GCK RRRNDIEFHVEITPWQVYGDAAGLGRAVLNLLDNAAKWSPPGGGVWVRLR
MSMEG_5487|M.smegmatis_MC2_155 RRRNDIEFDVDVIGWQVFGDAQGLGRAVLNLLDNAAKWSPPGGRVGVRLH
TH_2428|M.thermoresistible__bu RRRNDIDFDIEVVPWQVYGDAAGLSRAVMNLLDNAAKWSPSGGRVGVRLT
MLBr_00175|M.leprae_Br4923 RRRNDIHFDVQAIGWQIYGDAAGLSRAVLNLMDNAAKWSPSGGRVVVTMR
MAV_1095|M.avium_104 RRRNDIHFDVDVTPWQMYGDAAGLSRAVLNLLDNAAKWSPPGGHVGVTMR
Mb1008|M.bovis_AF2122/97 RRRNDIHFDVEVIGWQVYGDTAGLSRMALNLMDNAAKWSPPGGHVGVRLS
Rv0982|M.tuberculosis_H37Rv RRRNDILFDVEVIGWQVYGDTAGLSRMALNLMDNAAKWSPPGGHVGVRLS
MMAR_4528|M.marinum_M RRRNDIHFDVDVVGWQVYGDAAGLSRAALNLMDNAAKWSPPGGRVGIRLR
MUL_4700|M.ulcerans_Agy99 RRRNDIHFDVDVVGWQVYGDAAGLSRAALNLMDNAAKWSPPGGRVGIRLR
MAB_1077|M.abscessus_ATCC_1997 RRRNDIEFDVVMQPWQVYGDAAGLSRAVLNLLDNAAKWSPSGTAVSVRLA
****** *.: **::**: **.* .:**:********.* * : :
Mvan_4843|M.vanbaalenii_PYR-1 QIDALHAELVVSDRGPGIPPQERALVFERFFRSTSARSMPGSGLGLAIVK
Mflv_1888|M.gilvum_PYR-GCK QLDQLTAEVVVSDLGPGIPPAERELVFERFFRSTTARAMPGSGLGLAIVK
MSMEG_5487|M.smegmatis_MC2_155 QVDHMHAEIVVSDQGPGIPPEERRLVFERFYRSDAARAMPGSGLGLAIVQ
TH_2428|M.thermoresistible__bu QVDPVHAELVVSDHGPGIPPQERRLVFERFYRSTTARSMPGSGLGLAIVK
MLBr_00175|M.leprae_Br4923 QFDPSHVELVVSDYGPGIPPQERRLVFERFYRSTTARSLPGSGLGLAIVK
MAV_1095|M.avium_104 QLDPSHAELVVSDHGPGIPPQERRLVFERFYRSTSARAMPGSGLGLAIVK
Mb1008|M.bovis_AF2122/97 QLDASHAELVVSDRGPGIPVQERRLVFERFYRSASARALPGSGLGLAIVK
Rv0982|M.tuberculosis_H37Rv QLDASHAELVVSDRGPGIPVQERRLVFERFYRSASARALPGSGLGLAIVK
MMAR_4528|M.marinum_M QLDPSHAELVVSDNGPGISPQERRLVFERFYRSTSARAMPGSGLGLAIVK
MUL_4700|M.ulcerans_Agy99 QLDPSHAELVVSDNGPGISPQERRLVFERFYRSTSARAMPGSGLGLAIVK
MAB_1077|M.abscessus_ATCC_1997 QVDAAHAELVISDHGPGIPPAERDLVFERFYRATPARGMPGSGLGLAIVK
*.* .*:*:** ****. ** ******:*: .**.:**********:
Mvan_4843|M.vanbaalenii_PYR-1 QVVLKHGGTLRIEDTVP-----GGTPPGTAMHVVLPGRPSP---------
Mflv_1888|M.gilvum_PYR-GCK QVVLKHGGSLRVEDTVP-----GGEPPGTSVHVVLPGRPSA---------
MSMEG_5487|M.smegmatis_MC2_155 QVVLKHGGALRIDETVP-----GGNPPGASVHMLLPGQRIP---------
TH_2428|M.thermoresistible__bu QVVLKHGGTLRVEDTVP-----GGQPPGTSIHVVLPGRPLSGVPPASQEP
MLBr_00175|M.leprae_Br4923 QVVINHGGLLRVEDTAP-----GVQPPGTSIYVLLPGRPMPVSAYSTLAD
MAV_1095|M.avium_104 KVVLNHGGMLRVEDTVP-----GGQPPGTSFYVLLPGRPLPPAGHSTPAG
Mb1008|M.bovis_AF2122/97 QVVLNHGGLLRIEDTDP-----GGQPPGTSIYVLLPGRRMPIPQLPGAT-
Rv0982|M.tuberculosis_H37Rv QVVLNHGGLLRIEDTDP-----GGQPPGTSIYVLLPGRRMPIPQLPGAT-
MMAR_4528|M.marinum_M QVVLNHGGSLRIEDTVP-----GGQPPGTAICMLLPGRPMPDSAYPAAP-
MUL_4700|M.ulcerans_Agy99 QVVLNHGGSLRIEDTVP-----GGQPPGTAICMLLPGRPMPDSAYPAAP-
MAB_1077|M.abscessus_ATCC_1997 QVVLKHGGAIKVEETDPGAVIDGEPAPGTSMHVLLPGRPGPIVRP-----
:**::*** :::::* * * .**::. ::***: .
Mvan_4843|M.vanbaalenii_PYR-1 -------AGSDEAER-------------------------------
Mflv_1888|M.gilvum_PYR-GCK -------NLARGGE--------------------------------
MSMEG_5487|M.smegmatis_MC2_155 -----DPGATRSAEGFV------DDRGGHTVATE------------
TH_2428|M.thermoresistible__bu QPPQHAPGADPHAPTQT------GNDAVQSVFTAENK---------
MLBr_00175|M.leprae_Br4923 QDMGEANFQDKIGPAVQVSGKSANFRDSAHVISVDYQSARAR----
MAV_1095|M.avium_104 ESE-TDKAEAATDPAVPVAGDTANSRESANVISVDSQSARAR----
Mb1008|M.bovis_AF2122/97 -------AGARSTDIE-------NSRGSANVISVESQSTRAT----
Rv0982|M.tuberculosis_H37Rv -------AGARSTDIE-------NSRGSANVISVESQSTRAT----
MMAR_4528|M.marinum_M -------DDKKTEPVDTRGANGANSRGSANVISVDSQSARAR----
MUL_4700|M.ulcerans_Agy99 -------DDKKTEPVDTRGANGANSRGSANVISVDSQSARAR----
MAB_1077|M.abscessus_ATCC_1997 -------NMGDTAGRR-------NSGPNAREISVDSQSVPAAWRPS