For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SRFTEKMYRSAHTTGRGMVTGEPYEPVRHTWGEVHERAKRIAGGLAAAGVGLGDAVGVLAGFPVEIAPTA QGLWMRGASLTMLHQPTPRTDLVVWAEDTMNVIGMIEAKAVIVSEPFLVAIPVLEEKGIKVLTVTDLLAA DPIEPIEVGEDDLALMQLTSGSTGSPKAVQITHRNIYSNAEAMFIGAQYDVEKDVMVSWLPCFHDMGMVG FLTIPMYFGAELVKVTPLDFLRDTLLWAKLIDKYKGTMTAAPNFAYALFAKRLRRQAKPGEFDLSTLRFA LSGAEPVEPADVEDLLDAGKPFGLDPGAILPAYGMAETTLAVSFSPCGAGLTVDEVDADLLAALRRAVPA TKGHTKRLAELGPLLTDLEARVIDDHGNVMPPRGVGVIELRGECVTPGYMTMGGFIPAQDEHGWYDTGDL GYITEEGNVVVCGRVKDVIIMAGRNIYPTDIERAAGRVEGVRPGCAVAVRLDAGHSRETFAVAVESNNWQ DPAEVRRIEHQVAHEVVAEVDMRPRNVVVLGPGSIPKTSSGKLRRSNSVSLVT*
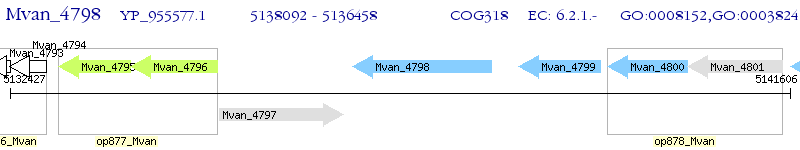
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_4798 | - | - | 100% (544) | acyl-CoA synthetase |
| M. vanbaalenii PYR-1 | Mvan_1051 | - | 0.0 | 72.98% (544) | acyl-CoA synthetase |
| M. vanbaalenii PYR-1 | Mvan_1945 | - | 4e-72 | 34.56% (515) | acyl-CoA synthetase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1041 | pks16 | 0.0 | 89.15% (544) | acyl-CoA synthetase |
| M. gilvum PYR-GCK | Mflv_1935 | - | 0.0 | 98.16% (544) | acyl-CoA synthetase |
| M. tuberculosis H37Rv | Rv1013 | pks16 | 0.0 | 89.15% (544) | acyl-CoA synthetase |
| M. leprae Br4923 | MLBr_00243 | - | 0.0 | 80.70% (544) | long-chain-fatty-acid--CoA ligase |
| M. abscessus ATCC 19977 | MAB_1140 | - | 0.0 | 75.55% (548) | acyl-CoA synthetase |
| M. marinum M | MMAR_4476 | pks16 | 0.0 | 82.17% (544) | polyketide synthase Pks16 |
| M. avium 104 | MAV_1150 | - | 0.0 | 90.99% (544) | acyl-CoA synthetase |
| M. smegmatis MC2 155 | MSMEG_5435 | - | 0.0 | 90.99% (544) | acyl-CoA synthetase |
| M. thermoresistible (build 8) | TH_3958 | - | 0.0 | 87.52% (537) | PUTATIVE AMP-dependent synthetase and ligase |
| M. ulcerans Agy99 | MUL_4648 | pks16 | 0.0 | 82.35% (544) | acyl-CoA synthetase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4476|M.marinum_M MSRFTDKMFRNARETTKGMVTGEPHNPVRHTWGEVHERARQIAGGLAAAG
MUL_4648|M.ulcerans_Agy99 MSRFTEKMFRNARETTKGMVTGEPHTPVRHTWGEVHERARQIAGGLAAAG
MLBr_00243|M.leprae_Br4923 MSKFTEKMFRNARESSKGIVTGEPHNPVRHTWGQVHERASRIAGGLAGVG
Mb1041|M.bovis_AF2122/97 MSRFTEKMFHNARTATTGMVTGEPHMPVRHTWGEVHERARCIAGGLAAAG
Rv1013|M.tuberculosis_H37Rv MSRFTEKMFHNARTATTGMVTGEPHMPVRHTWGEVHERARCIAGGLAAAG
MAV_1150|M.avium_104 MSRFTEKMYRNARTAKTGMVTGEPHNPVRHTWGEVHERARRIAGGLAAAG
Mvan_4798|M.vanbaalenii_PYR-1 MSRFTEKMYRSAHTTGRGMVTGEPYEPVRHTWGEVHERAKRIAGGLAAAG
Mflv_1935|M.gilvum_PYR-GCK MSRFTEKMYRSAHTTGRGMVTGEPYEPVRHTWSEVHERAKRIAGGLAAAG
MSMEG_5435|M.smegmatis_MC2_155 MSRFTEKMYRNARTVTTGMVTGEPHEPVRHTWGEVHERARRIAGGLAAAG
TH_3958|M.thermoresistible__bu -------MFHNARTATTGMVTGEPHAPVRHTWGEVHERARRIAGGLAAAG
MAB_1140|M.abscessus_ATCC_1997 MSRFTDELYANALSSSKGMVTGEPDTPVRHTWKQVHERAKQVAGGLAGAG
:: .* *:***** ****** :***** :*****..*
MMAR_4476|M.marinum_M VGPGDAVGVLAGAPVEIAPTAQGLWIRGASLTMLHQPTPRTDLALWAEDT
MUL_4648|M.ulcerans_Agy99 VGPGDAVGVLAGAPVEIAPTAQGLWIRGASLTMLHQPTPRTDLALWAEDT
MLBr_00243|M.leprae_Br4923 VGHGDAVGVLVGVPVEIAPTAQGLWMRGASLTMLHQPTPRTDLAVWEEDT
Mb1041|M.bovis_AF2122/97 VGLGDVVGVLAGFPVEIAPTAQALWMRGASLTMLHQPTPRTDLAVWAEDT
Rv1013|M.tuberculosis_H37Rv VGLGDVVGVLAGFPVEIAPTAQALWMRGASLTMLHQPTPRTDLAVWAEDT
MAV_1150|M.avium_104 IGPGDAVGVLAGFPVEIAPTAQGLWMRGASLTMLHQPTPRTDLAVWADDT
Mvan_4798|M.vanbaalenii_PYR-1 VGLGDAVGVLAGFPVEIAPTAQGLWMRGASLTMLHQPTPRTDLVVWAEDT
Mflv_1935|M.gilvum_PYR-GCK VGLGDAVGVLAGFPVEIAPTAQGLWMRGASLTMLHQPTPRTDLVMWAEDT
MSMEG_5435|M.smegmatis_MC2_155 IGHGDAVGVLAGFPVEIAPTAQGVWMRGASLTMLHQPTPRTDLAVWAEDT
TH_3958|M.thermoresistible__bu VGHGDAVGVLAGWPVEIAPTAQALWIRGASLTMLHQPTPRTDLMMWAEDT
MAB_1140|M.abscessus_ATCC_1997 LGPDDAIGMLVGMPVEIAPAIQAVWMRGASLTMLHQPTPRTDLAVWAEDT
:* .*.:*:*.* ******: *.:*:***************** :* :**
MMAR_4476|M.marinum_M TTVVDMIEAKAVVISDPFMAAAPVLEERGIRVLTFEQLLASEPIDPVEVG
MUL_4648|M.ulcerans_Agy99 TTVVDMIEAKAVVISDPFMAAAPVLEERGIRVLTFEQLLASEPIDPVEVG
MLBr_00243|M.leprae_Br4923 TTVIDMIEAKAVIISDPFMAVAPILEARGVKVVTVEQLLAADPVDPIETD
Mb1041|M.bovis_AF2122/97 MTVIGMIEAKAVIVSEPFLVAIPILEQKGMQVLTVADLLASDPIGPIEVG
Rv1013|M.tuberculosis_H37Rv MTVIGMIEAKAVIVSEPFLVAIPILEQKGMQVLTVADLLASDPIGPIEVG
MAV_1150|M.avium_104 MNVIGMIEAKAVVVSEPFLVAIPVLQEKDIKVLTVADLLASDPIDPVEVG
Mvan_4798|M.vanbaalenii_PYR-1 MNVIGMIEAKAVIVSEPFLVAIPVLEEKGIKVLTVTDLLAADPIEPIEVG
Mflv_1935|M.gilvum_PYR-GCK MNVIGMIEAKAVIVSEPFLVAIPVLEEKGIKVLTITDLLASEPIEPIEVG
MSMEG_5435|M.smegmatis_MC2_155 MNVIGMIELKAVIISEPFLVATPVLEEKGILVLKVADLLAADPIDPVETG
TH_3958|M.thermoresistible__bu MNVIGMIAAKVVVVSDPFTAAIPVLEERGIKVLTVEQLLAADPIDPVDTS
MAB_1140|M.abscessus_ATCC_1997 LKVVDMIGAKAVIVSAPFDAAAPVLEEKGVIVVHIKDLWDADPIEPLEAD
.*:.** *.*::* ** .. *:*: :.: *: . :* ::*: *::..
MMAR_4476|M.marinum_M EDELALMQLTSGSTGSPKAVQITHRNLFANLEAMYISAEYDAETDVMVSW
MUL_4648|M.ulcerans_Agy99 EDELALMQLTSGSTGSPKAVQITHRNLFANLEAMYISAEYDAETDVMVSW
MLBr_00243|M.leprae_Br4923 EDDLALMQLTSGSTGSPKAVQITHRNVYSNVEAMFIGVRYNVEKDVMISW
Mb1041|M.bovis_AF2122/97 EDDLALMQLTSGSTGSPKAVQITHRNIYSNAEAMFVGAQYDVDKDVMVSW
Rv1013|M.tuberculosis_H37Rv EDDLALMQLTSGSTGSPKAVQITHRNIYSNAEAMFVGAQYDVDKDVMVSW
MAV_1150|M.avium_104 EDDLALMQLTSGSTGSPKAVQITHRNIYSNAEAMFIGAQYDVDKDVMVSW
Mvan_4798|M.vanbaalenii_PYR-1 EDDLALMQLTSGSTGSPKAVQITHRNIYSNAEAMFIGAQYDVEKDVMVSW
Mflv_1935|M.gilvum_PYR-GCK EDDLALMQLTSGSTGSPKAVQITHRNIYSNAEAMFIGAQYDVEKDVMVSW
MSMEG_5435|M.smegmatis_MC2_155 EDDLALMQLTSGSTGSPKAVQITHRNIHSNAEAMFVGAKYDVEKDVMVSW
TH_3958|M.thermoresistible__bu EDDVALMQLTSGSTGSPKAVVITHRNIYSNAEAMFVGAQYDVDNDVMVSW
MAB_1140|M.abscessus_ATCC_1997 EDSVALMQLTSGSTGSPKAVKITHRNLYANAHGMYVASKYKPEIDVMVSW
**.:**************** *****:.:* ..*::. .*. : ***:**
MMAR_4476|M.marinum_M LPLFHDMGMIGFLTVPMYFAGELVKVTPMDFMRDTLLWAKLIDKYKGTMT
MUL_4648|M.ulcerans_Agy99 LPLFHDMGMIGFLTVPMYFAGELVKVTPMDFMRDTLLWAKLIDKYKGTMT
MLBr_00243|M.leprae_Br4923 LPCFHDMGMIGFLTVPMYFGAELVKVTPIDFLRDTLLWAKLIDKYKGTVI
Mb1041|M.bovis_AF2122/97 LPCFHDMGMVGFLTIPMFFGAELVKVTPMDFLRDTLLWAKLIDKYQGTMT
Rv1013|M.tuberculosis_H37Rv LPCFHDMGMVGFLTIPMFFGAELVKVTPMDFLRDTLLWAKLIDKYQGTMT
MAV_1150|M.avium_104 LPCFHDMGMVGFLTIPMYFGAELVKVTPMDFLRDTLLWAKLIDKYKGTMT
Mvan_4798|M.vanbaalenii_PYR-1 LPCFHDMGMVGFLTIPMYFGAELVKVTPLDFLRDTLLWAKLIDKYKGTMT
Mflv_1935|M.gilvum_PYR-GCK LPCFHDMGMVGFLTIPMYFGAELVKVTPMDFLRDTLLWAKLIDKYKGTMT
MSMEG_5435|M.smegmatis_MC2_155 LPCFHDMGMVGFLTIPMYFGAELVKVTPMDFLRDTLLWAKLIDKYKGTMT
TH_3958|M.thermoresistible__bu LPCFHDMGMIGFLTVPMYFGAELVKVTPMDFLRDTLLWARLIDKYKGTMT
MAB_1140|M.abscessus_ATCC_1997 LPLFHDMGMVGFLTVPMFFGAELVKITPMDFMRDVLLWGRLIDKYKGTMT
** ******:****:**:*..****:**:**:**.***.:*****:**:
MMAR_4476|M.marinum_M AAPNFAYSLFAKRLRKQAKPGQFDLSSLRFALSGAEPVEPSDVEDLCEAG
MUL_4648|M.ulcerans_Agy99 AAPNFAYSLFAKRLRKQAKPGQFDLSSLRFALSGAEPVEPSDVEDLCEAG
MLBr_00243|M.leprae_Br4923 CGPNFAYSLFTKRLRKQAKPGQFDLSTLRIAMSGAEPVDPADVEDLIDAG
Mb1041|M.bovis_AF2122/97 AAPNFAYALLAKRLRRQAKPGDFDLSTLRFALSGAEPVEPADVEDLLDAG
Rv1013|M.tuberculosis_H37Rv AAPNFAYALLAKRLRRQAKPGDFDLSTLRFALSGAEPVEPADVEDLLDAG
MAV_1150|M.avium_104 AAPNFAYALLAKRLRRQAKPGDFDLSTLRFALSGAEPVEPADVEDLLDAG
Mvan_4798|M.vanbaalenii_PYR-1 AAPNFAYALFAKRLRRQAKPGEFDLSTLRFALSGAEPVEPADVEDLLDAG
Mflv_1935|M.gilvum_PYR-GCK AAPNFAYALFAKRLRRQAKPGEFDLSTLRFALSGAEPVEPADVEDLLDAG
MSMEG_5435|M.smegmatis_MC2_155 AAPNFAYALLAKRLRRQAKPGDFDLSTLRFALSGAEPVEPADVEDFLDAG
TH_3958|M.thermoresistible__bu AAPNFAYALLAKRLRRQAKPGEFDLSTLRFALSGAEPVEPADVEDLIDAG
MAB_1140|M.abscessus_ATCC_1997 AAPNFAYSLFAKRLRNQAKPGDFDLSTLRFVLSGAEPVDPAVVEDLCDAG
..*****:*::****.*****:****:**:.:******:*: ***: :**
MMAR_4476|M.marinum_M APFGLRPQAILPAYGMAEITLAVAFSECGAG-LVVDEVDADLMAALRRAV
MUL_4648|M.ulcerans_Agy99 APFGLRPQAILPAYGMAEITLAVAFSECGAG-LVVDEVDADLMAALRRAV
MLBr_00243|M.leprae_Br4923 RPFGFRPEAILPAYGMAETTLAVSFSPVGEG-LLVDEVDADLLAALRRAV
Mb1041|M.bovis_AF2122/97 KPFGLRPSAILPAYGMAETTLAVSFSECNAG-LVVDEVDADLLAALRRAV
Rv1013|M.tuberculosis_H37Rv KPFGLRPSAILPAYGMAETTLAVSFSECNAG-LVVDEVDADLLAALRRAV
MAV_1150|M.avium_104 KPFGLKPSAILPAYGMAETTLAVSFSECNAG-LVVDEVDADLLAALRRAV
Mvan_4798|M.vanbaalenii_PYR-1 KPFGLDPGAILPAYGMAETTLAVSFSPCGAG-LTVDEVDADLLAALRRAV
Mflv_1935|M.gilvum_PYR-GCK KPFGLDPGAILPAYGMAETTLAVSFSPCGAG-LTVDEVDADLLAALRRAV
MSMEG_5435|M.smegmatis_MC2_155 KPFGLRPEAILPAYGMAETTLAVSFSPCGEG-LVVDEVDADLLAALRRAV
TH_3958|M.thermoresistible__bu KPFGLRPEAILPAYGMAETTLAVSFSECGAG-LIVDEVDADLLAALRRAV
MAB_1140|M.abscessus_ATCC_1997 KPFGFKPSAMVPAYGMAETALAVSFSEVEASGLVVDEVDADLLAALRRAV
***: * *::******* :***:** . * ********:*******
MMAR_4476|M.marinum_M PAAKGNTRRLASLGPLLQGIEARIVDADGNVLPSRGVGVIELRGESVTPG
MUL_4648|M.ulcerans_Agy99 PAAKGNTRRLASLGPLLQGIEARIVDADGNVLPSRGVGVIELRGESVTPG
MLBr_00243|M.leprae_Br4923 PATKGNIRRLVSLGPLLDGLEVRIVDEDGNVLPSRGVGVIELRGESLTPG
Mb1041|M.bovis_AF2122/97 PATKGNTRRLATLGPLLQDLEARIIDEQGDVMPARGVGVIELRGESLTPG
Rv1013|M.tuberculosis_H37Rv PATKGNTRRLATLGPLLQDLEARIIDEQGDVMPARGVGVIELRGESLTPG
MAV_1150|M.avium_104 PATKGNTRRLATLGPLLQDLEARIVDENGDVMPPRGVGVIELRGESVTPG
Mvan_4798|M.vanbaalenii_PYR-1 PATKGHTKRLAELGPLLTDLEARVIDDHGNVMPPRGVGVIELRGECVTPG
Mflv_1935|M.gilvum_PYR-GCK PASKGHTKRLAELGPLLTDLEARVIDDHGNLMPPRGVGVIELRGECVTPG
MSMEG_5435|M.smegmatis_MC2_155 PATKGNTKRLASLGPLLNDLEARVVDENGEVMPARGVGVIELRGEPVTPG
TH_3958|M.thermoresistible__bu PASKGNTRRLASLGPLLPDLEARVVDEDGNILPPRGVGIIELRGEPVTPG
MAB_1140|M.abscessus_ATCC_1997 PATRGNLRHLATLGPLLPGIEARVVDEDLNVLPARGVGVIELRGESVTPG
**::*: ::*. ***** .:*.*::* . :::*.****:****** :***
MMAR_4476|M.marinum_M YLTMGGFIPAQDQLGWYDTGDLGYQMENGHVVVCGRVKDVIIMAGRNIYP
MUL_4648|M.ulcerans_Agy99 YLTMGGFIPAQDQLGWYDTGDLGYQMENGHVVVCGRVKDVIIMAGRNIYP
MLBr_00243|M.leprae_Br4923 YITMGGFISAQDEHGWYDTGDLGYQMENGHIVVCGRVKDVIIMAGRNVYP
Mb1041|M.bovis_AF2122/97 YLTMGGFIPAQDEHGWYDTGDLGYLTEEGHVVVCGRVKDVIIMAGRNIYP
Rv1013|M.tuberculosis_H37Rv YLTMGGFIPAQDEHGWYDTGDLGYLTEEGHVVVCGRVKDVIIMAGRNIYP
MAV_1150|M.avium_104 YLTMGGFIPAQDENGWYDTGDLGYLTEEGHVVVCGRVKDVIIMAGRNIYP
Mvan_4798|M.vanbaalenii_PYR-1 YMTMGGFIPAQDEHGWYDTGDLGYITEEGNVVVCGRVKDVIIMAGRNIYP
Mflv_1935|M.gilvum_PYR-GCK YMTMGGFIPAQDEHGWYDTGDLGYITEEGNVVVCGRVKDVIIMAGRNIYP
MSMEG_5435|M.smegmatis_MC2_155 YITMGGFVPAQDEYGWYDTGDLGYITEEGNIVVCGRVKDVIIMAGRNIYP
TH_3958|M.thermoresistible__bu YVTMGGFVPAQDEHGWYDTGDLGYLTEEGHVVVCGRVKDVIIMAGRNIYP
MAB_1140|M.abscessus_ATCC_1997 YVTMGGFLAAQDENGWYNTGDLGYLTEKGHVVVCGRVKDVIIMAGRNIYP
*:*****:.***: ***:****** *:*::****************:**
MMAR_4476|M.marinum_M TDIERAAGRVEGVRPGCAVAVRLDAGHS----RETFAVAVESNAFEDPAE
MUL_4648|M.ulcerans_Agy99 TDIERAAGRVEGVRPGCAVAVRLDAGHS----RETFAVAVESNAFEDPAE
MLBr_00243|M.leprae_Br4923 TDIERAAGRVDGVRPGCAVAVRLDAGHS----RETFAVAVESNAFEDTAE
Mb1041|M.bovis_AF2122/97 TDIERAAGRVDGVRPGCAVAVRLDAGHS----RESFAVAVESNAFEDPAE
Rv1013|M.tuberculosis_H37Rv TDIERAAGRVDGVRPGCAVAVRLDAGHS----RESFAVAVESNAFEDPAE
MAV_1150|M.avium_104 TDIERAACRVEGVRPGCAVAVRLDAGHS----RETFAVAVESNAFQDPAE
Mvan_4798|M.vanbaalenii_PYR-1 TDIERAAGRVEGVRPGCAVAVRLDAGHS----RETFAVAVESNNWQDPAE
Mflv_1935|M.gilvum_PYR-GCK TDIERAAGRVEGVRPGCAVAVRLDAGHS----RETFAVAVESNNWQDPVE
MSMEG_5435|M.smegmatis_MC2_155 TDIERAAGRVEGVRPGCAVAVRLDAGHS----RETFAVAVESNAWQDPAE
TH_3958|M.thermoresistible__bu TDIERAAQRVEGVRPGGAVAVRLDAGHS----RETFAVAVESNAWQDPEE
MAB_1140|M.abscessus_ATCC_1997 TDIERAAGRVDGVRPGCAVAVRLEAGVDPKNQRETFAVVVESNAFENPDE
******* **:***** ******:** . **:***.**** :::. *
MMAR_4476|M.marinum_M VRRIEHQVAHEVVAEVDVRPRNVVVLGPGTIPKTPSGKLRRSNSLTLVT-
MUL_4648|M.ulcerans_Agy99 VRRIEHQVAHEVVAEVDVRPRNVVVLGPGTIPKTPSGKLRRSNSLTLVT-
MLBr_00243|M.leprae_Br4923 VRRIEHQVAHEVLKEVDVRPRNVVVLGPGTIPKTPSGKLRRASSVTLVT-
Mb1041|M.bovis_AF2122/97 VRRIEHQVAHEVVAEVDVRPRNVVVLGPGTIPKTPSGKLRRANSVTLVT-
Rv1013|M.tuberculosis_H37Rv VRRIEHQVAHEVVAEVDVRPRNVVVLGPGTIPKTPSGKLRRANSVTLVT-
MAV_1150|M.avium_104 VRRIEHQVAREVVAEVDMRPRNVVVLGPGTIPKTPSGKLRRSNSVTLVT-
Mvan_4798|M.vanbaalenii_PYR-1 VRRIEHQVAHEVVAEVDMRPRNVVVLGPGSIPKTSSGKLRRSNSVSLVT-
Mflv_1935|M.gilvum_PYR-GCK VRRIEHQVAHEVVAEVDVRPRNVVVLGPGSIPKTSSGKLRRSNSVSLVT-
MSMEG_5435|M.smegmatis_MC2_155 VRRIEHQVAHEVVSEVDVRPRNVVVLGPGSIPKTPSGKLRRANSVSLVTQ
TH_3958|M.thermoresistible__bu VRRIEHQVAHEVVAEVDVRPRNVVVLGPGSIPKTSSGKLRRSTSVSLVT-
MAB_1140|M.abscessus_ATCC_1997 VRRIEQQVAHEVVAEVDVRPRNVVVLGPGSIPKTPSGKLRRSTSVALVI-
*****:***:**: ***:***********:****.******:.*::**