For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
GAWKNATAGEPLRGVVGRPASSSVFDLRRWAADGRTGRYVEHFWSVAWDLRGRDPFDTSVITFPAMHITR EWGEDGARHGFPLPNTLVHGVVEQVFRTTISERGWVVGARFRPGGFTARFGGDASTMTGRIQPVDDLSDV FGGPVLVPDDVDAAAAEFDRLIAHGAEPDDTYRALAAVVDRIRDDSGIHRVEQVMSHSPWSVRTTQRVFR RYVGVPVKWVLCRYRLQNAALEIETDPAVDFADLAVRLGWYDQAHFINDFRAMLGSTPGEYAATHGRRP*
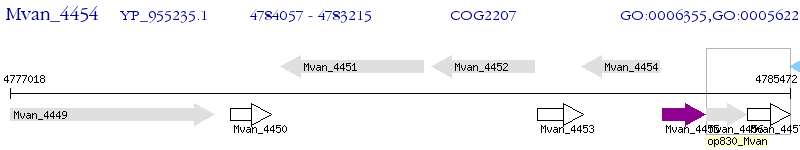
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_4454 | - | - | 100% (280) | helix-turn-helix domain-containing protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_2241 | - | 1e-133 | 80.22% (278) | helix-turn-helix domain-containing protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0738 | - | 4e-30 | 33.92% (286) | AraC family transcriptional regulator |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_0221 | - | 1e-09 | 28.18% (220) | transcriptional regulator, AraC family protein |
| M. smegmatis MC2 155 | MSMEG_5025 | - | 1e-108 | 67.14% (280) | putative transcriptional regulator |
| M. thermoresistible (build 8) | TH_1844 | - | 1e-102 | 62.77% (282) | putative transcriptional regulator |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_4454|M.vanbaalenii_PYR-1 --MGAWKNATA------GEPLRGVVGRPASSSVFDLRRWAADGRTGRYVE
Mflv_2241|M.gilvum_PYR-GCK --MAAWKNTTA------GAPLRGVVGRPAGASAFDLRRWAADGRTSRYVE
MSMEG_5025|M.smegmatis_MC2_155 --MAAWKN----------APERGVVGRPGSASAFDLTRWAPSPEAGRFVE
TH_1844|M.thermoresistible__bu VTVGAWKNASPP-----VAPVRGVVGRAVDASVYDLYRWEPSAEAAPFVE
MAB_0738|M.abscessus_ATCC_1997 --MPSWTNPTEPREGLGADPWRAVVRPEGLLPRIDRVVLPGRGPLRDWVE
MAV_0221|M.avium_104 -------------------------------------MYRPSPPLAEHIE
:*
Mvan_4454|M.vanbaalenii_PYR-1 HFWSVAWDLRGRDPFDTSVITFPAMHITREWGEDG-------ARHGFPLP
Mflv_2241|M.gilvum_PYR-GCK HYWSVNWDLRGNDPFDSTVITFPAMHITHEWGQDA-------VRHGFSLP
MSMEG_5025|M.smegmatis_MC2_155 HYWSVRWDRREAGPHESTVITFPAVHLTREWGDDI-------ARHGHPLP
TH_1844|M.thermoresistible__bu HFWAVSWDLRDRPPQPSTVISFPVMHLTHEWGEDG-------VRHGFSLP
MAB_0738|M.abscessus_ATCC_1997 RIWSLSWAEPVPADFSG-VIAHPTVHLTLEGGPPGE------IRHGHRLP
MAV_0221|M.avium_104 YFGYWRGDEALGVHTSRALPRGAVTAIIDVGGRTDLGFYASDARTPLTVP
: .. : * * :*
Mvan_4454|M.vanbaalenii_PYR-1 NTLVHGVVEQVFRTTISERGWVVGARFRPGGFTARFGGDASTMTGRIQPV
Mflv_2241|M.gilvum_PYR-GCK NTLVHGVVEQVFRTTISEKGCVVGARFRPGGFTARFGGDASVRTGCVEPI
MSMEG_5025|M.smegmatis_MC2_155 NTLLHGVVTQVFRTTISSRGVVVGARFHPGGFTARFGRDAGELTDRVVPV
TH_1844|M.thermoresistible__bu ATLIHGVVPQVFRITLNERGAVAGARFRPGGFTARFGRDAASYTGRVVRA
MAB_0738|M.abscessus_ATCC_1997 AGLLHGVVTERFAVDLPAPGWVVGLHLRPGAIFDLTGTEASTWTGRVVPW
MAV_0221|M.avium_104 PAFAAGAGATAYVVRVAPAHTVMTIHFRPAGALAFLGSPLSDLEDALVGL
: *. : : * :::*.. * . . :
Mvan_4454|M.vanbaalenii_PYR-1 DDLS--D-VFGGPVLVPDDVDAAAAEFDRLIAH-GAEPDDTYR--ALAAV
Mflv_2241|M.gilvum_PYR-GCK GDL------FGAAVALPDDVEDAAAELDTLIGG-GGPPDAVFD--ALSAL
MSMEG_5025|M.smegmatis_MC2_155 DDD-----LFGGPIDLGCDVSVVRSGFDAVIGDYSGDPDPVYA--RLMPL
TH_1844|M.thermoresistible__bu EHEL--SGELSIGTIISDELTAVAEQLDTVIGGHRGAPDTTYV--ALTRL
MAB_0738|M.abscessus_ATCC_1997 DQAW--PGWDLTAVWEACDARARAEAIERAAQEMLGGRRPTADGHRARSV
MAV_0221|M.avium_104 EDLWGRDAALLREQLIDAGSPPRRVALLQAFLVRRMRRNAVWPPARLAPV
. : . :
Mvan_4454|M.vanbaalenii_PYR-1 VDRIRDDSGIHRVEQVMSHSPWSVRTTQRVFRRYVGVPVKWVLCRYRLQN
Mflv_2241|M.gilvum_PYR-GCK VDRIRDDSGVHRVEQVMDLSPWSVRTTQRVFRRYVGVPVKWVLCRYRLQN
MSMEG_5025|M.smegmatis_MC2_155 LERMREDPTLHRVEQVMEHSPWSVRTTQRVFRRYVGVPVKWVLCRYRLQQ
TH_1844|M.thermoresistible__bu LDRIREDDRMHRVDQVMEHAPWSVRTTQRIFRRYVGVPVKWVLCRYRLQQ
MAB_0738|M.abscessus_ATCC_1997 ERLVRTDPSIASVADLAARAALSQRSLQRLCRSHLGVSPRWLLRRARVID
MAV_0221|M.avium_104 LRGADLDPSMR-VSKAQELSGLSRKRFAALFRCEVGLSPKAYLRVLRLQA
* : * . : * : : * :*:. : * *:
Mvan_4454|M.vanbaalenii_PYR-1 AALEIETDPAVDFADLAVRLGWYDQAHFINDFRAMLGSTPGEYAATHGRR
Mflv_2241|M.gilvum_PYR-GCK AALEIETDPGVDFADLAVRLGWYDQAHFINDFRSMLGSTPGEYAATYGQG
MSMEG_5025|M.smegmatis_MC2_155 AALEIESTGSVDFADLAVRLGWYDQAHFINDFRGMLGCTPGEYAMRFGQK
TH_1844|M.thermoresistible__bu AALEIESSPDVDFADLAVRLGWYDQPHFINDFRAMLGCTPAQYADRYRSR
MAB_0738|M.abscessus_ATCC_1997 AHQLLS-ETDLDVAEVASRLGWYDQAHLTRDYTKLTGTPPVRLRQERREG
MAV_0221|M.avium_104 ALRALDTPARG--ATIAADLGYFDQAHFVREFRAFTGATPTQYARRRSSM
* :. * :* **::**.*: .:: : * .* .
Mvan_4454|M.vanbaalenii_PYR-1 P-------
Mflv_2241|M.gilvum_PYR-GCK R-------
MSMEG_5025|M.smegmatis_MC2_155 GGGS----
TH_1844|M.thermoresistible__bu T-------
MAB_0738|M.abscessus_ATCC_1997 R-------
MAV_0221|M.avium_104 PGHVELAR