For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MRSVALLVLFCFLASASVVSAQAEPPGIPDLRGYRTVPVDDFVVDGAAYFQTPDGLDCAILPTNGTAGCD GPLPATPAGANEIVLAAEVDTRGLRTTANPSFLAPPGHAVPELPEGAKIVYGDFECAAGSGPITLCAKGT PAMQWMMISAERTGIGPATDGLPPGFPDPNDFVVGDDEYLVGTGPKNMFPIFTVEGGLTCSIVTFSGGEI GCNGPLPGVSGGENEIFAQLPGATGIRRTDNPKFSTPDYPGQIRQLPVGHRVNGIGGTCMAIAGGVACYG TVAGRAQGFEVSATETTTFG
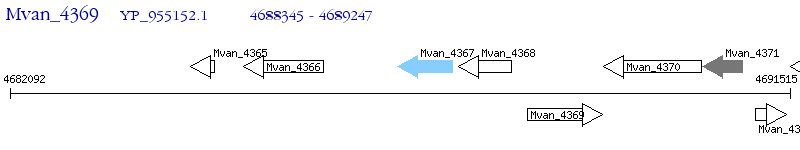
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_4369 | - | - | 100% (300) | hypothetical protein Mvan_4369 |
| M. vanbaalenii PYR-1 | Mvan_0994 | - | 1e-09 | 34.51% (113) | hypothetical protein Mvan_0994 |
| M. vanbaalenii PYR-1 | Mvan_2552 | - | 4e-06 | 23.90% (272) | hypothetical protein Mvan_2552 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_5250 | - | 7e-09 | 34.19% (117) | hypothetical protein Mflv_5250 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3917c | - | 5e-11 | 30.66% (137) | hypothetical protein MAB_3917c |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_0501 | - | 7e-09 | 31.88% (138) | hypothetical protein MAV_0501 |
| M. smegmatis MC2 155 | MSMEG_1136 | - | 3e-09 | 29.48% (173) | hypothetical protein MSMEG_1136 |
| M. thermoresistible (build 8) | TH_1765 | - | 1e-10 | 34.43% (122) | conserved hypothetical protein |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_1136|M.smegmatis_MC2_155 -MLSRLLVGLASAAGAAVMSVPGVAAADPQADPPPPTV--PDVNAYPPVK
TH_1765|M.thermoresistible__bu -VLKRVLLG---AALAAVVGVPAVAAAEP--DRPGP----PDVNAFEPVR
Mflv_5250|M.gilvum_PYR-GCK MVLSRSVVAAVCVAVAGVLNASGLAAAQPPPPPPPPPA--PDINAYPPAN
MAB_3917c|M.abscessus_ATCC_199 -MRILPILGAVASAVVAVM-LAGVASADPDTPNPLDPSGLPNVNGLTPVS
MAV_0501|M.avium_104 ------MLSKTFNATAAVIGTAVISACTAHAEPPKF----PDLDAFQPVD
Mvan_4369|M.vanbaalenii_PYR-1 -----------MRSVALLVLFCFLASASVVSAQAEPPG-IPDLRGYRTVP
: . :: :::. . *:: . ..
MSMEG_1136|M.smegmatis_MC2_155 TSEYAVMDNN--WYAFSIGEGITCVLQRT------GSYGCSGPIPAAPNG
TH_1765|M.thermoresistible__bu LSEYTVMDGA--WFAFRGPAGITCVLQRS------GGYGCSGPLPGAPGG
Mflv_5250|M.gilvum_PYR-GCK PKDFAVHQGS--AYAFTT-AGLTCMVQRS------GPYGCSGVLPGAPNN
MAB_3917c|M.abscessus_ATCC_199 PLEYSTLADT--AYGFTIPGGISCLIKRAD-----ASYGCSGPLPGAPDG
MAV_0501|M.avium_104 PAPYTASGRAGGGAYFVTPDGIQCSVPHPDHPGEHVSVSCAGPLPGLPDD
Mvan_4369|M.vanbaalenii_PYR-1 VDDFVVDGAA----YFQTPDGLDCAILPTN-----GTAGCDGPLPATPAG
: . * *: * : . .* * :*. * .
MSMEG_1136|M.smegmatis_MC2_155 AN-LVSGGPG-VPGFASSPAPVFAVVGD---------AKPLPPNTRLSYQ
TH_1765|M.thermoresistible__bu AN-MVSGGPG-VPGFATTDASVFDVVGD---------AEPLPPNSRISYQ
Mflv_5250|M.gilvum_PYR-GCK AN-LVSGRSGTVPGFGAAPQPIVGEP-----------VNALPPNTRISFG
MAB_3917c|M.abscessus_ATCC_199 AN-LVSGSNA--PGFATTDRPIYGMGGEP--------FKPLPPGSRLSYR
MAV_0501|M.avium_104 AP-RGKDGCSAVGSPTSLPTDLGPYSFQEGTGCPILTTPPLNVGQKITTA
Mvan_4369|M.vanbaalenii_PYR-1 ANEIVLAAEVDTRGLRTTANPSFLAPPG-------HAVPELPEGAKIVYG
* . : * . ::
MSMEG_1136|M.smegmatis_MC2_155 TISCGFD-GVTTACIDGRNQAGFVISPTGSFVIGQQESDFKPTFQIG---
TH_1765|M.thermoresistible__bu TVSCGTD-GTMTTCVDSRTGAGFVIGPAGSHILAPSNPLLNRRN------
Mflv_5250|M.gilvum_PYR-GCK TVSCGGD-GTVTACVDSMNQAGFVVSPDATWIVNAVNPLVARPEGTNPYF
MAB_3917c|M.abscessus_ATCC_199 EVSCGLDSGGTLTCVNNRWQNGFVVGPGGSYTT-----------------
MAV_0501|M.avium_104 GITCAVGGDRLTACIDPVLNRGFVLEPSGSWTF-----------------
Mvan_4369|M.vanbaalenii_PYR-1 DFECAAGSGPITLCAKGTPAMQWMMISAERTGIGPATDGLPPGFPDPNDF
. *. . . * . ::: .
MSMEG_1136|M.smegmatis_MC2_155 --------------------------------------------------
TH_1765|M.thermoresistible__bu --------------------------------------------------
Mflv_5250|M.gilvum_PYR-GCK N-------------------------------------------------
MAB_3917c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_0501|M.avium_104 --------------------------------------------------
Mvan_4369|M.vanbaalenii_PYR-1 VVGDDEYLVGTGPKNMFPIFTVEGGLTCSIVTFSGGEIGCNGPLPGVSGG
MSMEG_1136|M.smegmatis_MC2_155 --------------------------------------------------
TH_1765|M.thermoresistible__bu --------------------------------------------------
Mflv_5250|M.gilvum_PYR-GCK --------------------------------------------------
MAB_3917c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_0501|M.avium_104 --------------------------------------------------
Mvan_4369|M.vanbaalenii_PYR-1 ENEIFAQLPGATGIRRTDNPKFSTPDYPGQIRQLPVGHRVNGIGGTCMAI
MSMEG_1136|M.smegmatis_MC2_155 ----------------------------
TH_1765|M.thermoresistible__bu ----------------------------
Mflv_5250|M.gilvum_PYR-GCK ----------------------------
MAB_3917c|M.abscessus_ATCC_199 ----------------------------
MAV_0501|M.avium_104 ----------------------------
Mvan_4369|M.vanbaalenii_PYR-1 AGGVACYGTVAGRAQGFEVSATETTTFG