For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TRPGPRISPALAGAVDLSALKQRASAGGGAPAGNPGGVEVTEANLEAEVLVRSTEVPVVVLLWSPRSDSS VQLGDALAALSAADGAKWTFATVNVDTTPRVAQMFGVQAVPTVVALAAGRPLSSFEGIQPPEQLRRWIDS LLNAVAGKLGGTGEDEQAEAVDPEVEQARAHLDAGDFEAAQSAYQAILDANPNHDEAKGAVRQIGFLQRA TAHPRDAIAVADASPNDIDAAFAAADVEILQQDVQAAFTRLTALVKRTAGDDRTKVRTRLIELFDLFDPA DPEVVAGRRNLANALY*
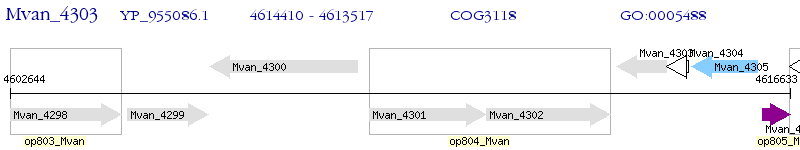
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_4303 | - | - | 100% (297) | thioredoxin domain-containing protein |
| M. vanbaalenii PYR-1 | Mvan_5905 | - | 3e-10 | 38.54% (96) | thioredoxin |
| M. vanbaalenii PYR-1 | Mvan_6068 | - | 4e-05 | 24.76% (105) | thioredoxin |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1359 | - | 1e-106 | 65.79% (304) | thioredoxin |
| M. gilvum PYR-GCK | Mflv_2342 | - | 1e-137 | 82.55% (298) | thioredoxin domain-containing protein |
| M. tuberculosis H37Rv | Rv1324 | - | 1e-106 | 65.79% (304) | thioredoxin |
| M. leprae Br4923 | MLBr_01159 | - | 1e-96 | 60.80% (301) | hypothetical protein MLBr_01159 |
| M. abscessus ATCC 19977 | MAB_1465 | - | 1e-91 | 59.61% (307) | hypothetical protein MAB_1465 |
| M. marinum M | MMAR_4074 | - | 1e-112 | 68.75% (304) | thioredoxin |
| M. avium 104 | MAV_1545 | - | 1e-115 | 71.29% (303) | thioredoxin domain-containing protein |
| M. smegmatis MC2 155 | MSMEG_4917 | - | 1e-121 | 74.16% (298) | TPR repeat-containing protein |
| M. thermoresistible (build 8) | TH_1534 | - | 1e-124 | 75.17% (298) | POSSIBLE THIOREDOXIN |
| M. ulcerans Agy99 | MUL_3937 | - | 1e-112 | 68.42% (304) | thioredoxin |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1359|M.bovis_AF2122/97 MTRPRPPLG--PAMAGAVDLSGIKQRAQQNAAASTDADRALSTPSG----
Rv1324|M.tuberculosis_H37Rv MTRPRPPLG--PAMAGAVDLSGIKQRAQQNAAASTDADRALSTPSG----
MMAR_4074|M.marinum_M MTRPRPPIG--PALAGAVDLSGLKQRAQQDTAAGGASGKAPADQPG----
MUL_3937|M.ulcerans_Agy99 MTRPRPPIG--PALAGAVDLSGLKQRAQQDTAAGGASGKAPADQPG----
MLBr_01159|M.leprae_Br4923 -------------MAGAVDLSGLKQRARQKASTSDPASRATLGARGTGSS
MAV_1545|M.avium_104 MTRPRPSIG--PALAGAVDLSGLKQRAQAASGPAGGPAPADGGPG-----
Mvan_4303|M.vanbaalenii_PYR-1 MTRPGPRIS--PALAGAVDLSALKQR-ASAGGGAPAGNPGG---------
Mflv_2342|M.gilvum_PYR-GCK MTRPGPRIS--SALAGAVDLSALKQRPASSAESASTGAPGG---------
MSMEG_4917|M.smegmatis_MC2_155 MTRPRPSIG--PALAGAVDLSALKRPAASGEAAPAPGGPNV---------
TH_1534|M.thermoresistible__bu VTRPGPSIG--PALAGAVDLSALKQRSAANQEGTGSAPPGG---------
MAB_1465|M.abscessus_ATCC_1997 MTRPRPAVAAAAAMSGAVDLSALKQRAQEPASGEPSAGGEWT--------
::******.:*:
Mb1359|M.bovis_AF2122/97 ----VTEITEANFEDEVIVRSDEVPVVVLLWSPRSEVCVDLLDTLSGLAA
Rv1324|M.tuberculosis_H37Rv ----VTEITEANFEDEVIVRSDEVPVVVLLWSPRSEVCVDLLDTLSGLAA
MMAR_4074|M.marinum_M ----VVEVTEANFEDEVIGRSDEVPVVVVLWSPRSDVCVELVDTLSGLAA
MUL_3937|M.ulcerans_Agy99 ----VVEVTEANFEDEVIGRSDEVPVVVVLWSPRSDVCVELVDTLSGLAA
MLBr_01159|M.leprae_Br4923 ENTSVIEITEANFEDEVLVRSHEVPILVLVWSPRSDACVKLLETLSGLAV
MAV_1545|M.avium_104 ----ISAVTEANFEAEVLIRSEEVPVVVLLWSPRSDACVQLLDTLSGLAA
Mvan_4303|M.vanbaalenii_PYR-1 -----VEVTEANLEAEVLVRSTEVPVVVLLWSPRSDSSVQLGDALAALSA
Mflv_2342|M.gilvum_PYR-GCK -----IEVTETNLEADVLVRSTEVPVVVLLWSPRSDASVQLGEALGALSA
MSMEG_4917|M.smegmatis_MC2_155 -----TEVTEANFEAEVLVRSGQVPVVVVLWSPRSDASAQLTETFADLAS
TH_1534|M.thermoresistible__bu -----VEVTETNLEAEVLVRSTQVPVVVLLWTPRSDASVQLGNTLGTLAS
MAB_1465|M.abscessus_ATCC_1997 -----VEVTEANLESEVLAQSNRVPVIVLLGSPRSEASAALAATLGDLVA
:**:*:* :*: :* .**::*:: :***: .. * ::. *
Mb1359|M.bovis_AF2122/97 AAKGKWSLASVNVDVAPRVAQIFGVQAVPTVVALAAGQPISSFQGLQPAD
Rv1324|M.tuberculosis_H37Rv AAKGKWSLASVNVDVAPRVAQIFGVQAVPTVVALAAGQPISSFQGLQPAD
MMAR_4074|M.marinum_M ADMGKWRLATVNVDVVPRVAQIFGVQAVPTVVALAAAQPISSFQGPQPAE
MUL_3937|M.ulcerans_Agy99 ADMGKWRLATVNVDVVPRVAQIFGVQAVPTVVALAAAQPISSFQGPQPAE
MLBr_01159|M.leprae_Br4923 ADSGTWLLATVNVDAVPRVAQIFGVDAVPTVVALAAGQPLSSFQGMQSVD
MAV_1545|M.avium_104 QDNGKWSLATVNVDVAPRVAQIFGVDAVPTVVALAAGQPISSFQGMQPAD
Mvan_4303|M.vanbaalenii_PYR-1 ADGAKWTFATVNVDTTPRVAQMFGVQAVPTVVALAAGRPLSSFEGIQPPE
Mflv_2342|M.gilvum_PYR-GCK TDGAKWVFATVNVDTTPRVAQMFGVQAVPTVVALAAGRPLSSFEGMQPPD
MSMEG_4917|M.smegmatis_MC2_155 ADGGKWSLAAVNVDTTPRIAQMFGIQAVPTVVALAGGQPIASFQGPQPPE
TH_1534|M.thermoresistible__bu EDGGKWAFATVNVDTTPRVAQMFGVQAVPTVVALAGGQPISSFQGVQPAD
MAB_1465|M.abscessus_ATCC_1997 EDRGTWALARVNVDTNPQIAQVFGVQAVPTVVAVAAGRPLTSFTGPQPAD
..* :* ****. *::**:**::*******:*..:*::** * *. :
Mb1359|M.bovis_AF2122/97 QLSRWVDSLLSATAGKLKGAASSEE---STEVDPAVAQARQQLEDGDFVA
Rv1324|M.tuberculosis_H37Rv QLSRWVDSLLSATAGKLKGAASSEE---STEVDPAVAQARQQLEDGDFVA
MMAR_4074|M.marinum_M QLRLWVDSLLSATDGKLSGVAGSED---AEEVDPTVAQARAELEEGDFDA
MUL_3937|M.ulcerans_Agy99 QLRLWVDSLLSATDGKLSGVAGSED---AEEVDPTVAQARAELEEGNFDA
MLBr_01159|M.leprae_Br4923 QLRRWLDSLLSVTAGKLRGPTRSED---SAEIDPAIAQARQQLEAGDFLT
MAV_1545|M.avium_104 QLRGWLDQILSATAGKLKGATGSAE---SEAVDPELAAARQQLEAGDFEA
Mvan_4303|M.vanbaalenii_PYR-1 QLRRWIDSLLNAVAGKLGGTGEDEQ---AEAVDPEVEQARAHLDAGDFEA
Mflv_2342|M.gilvum_PYR-GCK QLRRWIDSLLNAVAGKLSGAGDEGQ---PEEVDPQLEQARALLDEGDFDA
MSMEG_4917|M.smegmatis_MC2_155 QLRRWVDSLLEATAGKLSGS-GEE----AEQVDPALAQARAHLDQGNFDE
TH_1534|M.thermoresistible__bu QLRRWIDSLLNATAGKLSGPPGEEQ---AEQVDPEVAQARAQLDAGDFDG
MAB_1465|M.abscessus_ATCC_1997 QLRRWLDSILDAVAGKLPDAPVSDDEDTEEAVDPALAAARDALDSGDFEA
** *:*.:*... *** . . :** : ** *: *:*
Mb1359|M.bovis_AF2122/97 ARKSYQAILDANPG---SVEAKAAIRQIEFLIRATAQRPDAVSVADSLSD
Rv1324|M.tuberculosis_H37Rv ARKSYQAILDANPG---SVEAKAAIRQIEFLIRATAQRPDAVSVADSLSD
MMAR_4074|M.marinum_M ARGSYQAVLDANPN---SVEAKAAIRQIDFLSRATAQRPDAVLVADAAPD
MUL_3937|M.ulcerans_Agy99 ARGSYQAVLDANPN---SVEAKAAIRQIDFLSRATAQRPDAVLVADAAPD
MLBr_01159|M.leprae_Br4923 AKQSYHAILDADPA---SVEAKAAIRQIDFLTRATAQHPDAVAVADAAPG
MAV_1545|M.avium_104 ARTSYQAILDANPA---SAEAKGAIRQIDFLTRATAQRPDAVAVADAAPG
Mvan_4303|M.vanbaalenii_PYR-1 AQSAYQAILDANPN---HDEAKGAVRQIGFLQRATAHPRDAIAVADASPN
Mflv_2342|M.gilvum_PYR-GCK ARNAYQAILDADPN---HAEAKGAVRQIGFLQRATTHPQDAVAIADAAPD
MSMEG_4917|M.smegmatis_MC2_155 ALKAYEAILEAQPN---HPEAKGAVRQIGFLQRATAQRPDAVEVADAAPD
TH_1534|M.thermoresistible__bu ALKSYKAILETRPN---HPEAKGAVRQIEFLQRATKHSPDAIATADAAPD
MAB_1465|M.abscessus_ATCC_1997 ARTAYQGLLDGGATGAVAAEATASVRQIAFLIRATGQPQDAVLTANAAPG
* :*..:*: . **..::*** ** *** : **: *:: ..
Mb1359|M.bovis_AF2122/97 DIDAAFAAADVQVLNQDVSAAFERLIALVRRTSGEERTRVRTRLIELFEL
Rv1324|M.tuberculosis_H37Rv DIDAAFAAADVQVLNQDVSAAFERLIALVRRTSGEERTRVRTRLIELFEL
MMAR_4074|M.marinum_M DVDAAFAAADVQVLSQDVTGAFERLVALVRRTSGDERTRVRTRLIELFEL
MUL_3937|M.ulcerans_Agy99 DVDAAFAAADVQVLSQDVTGAFERLVALVRRTSGDERTRVRTRLIELFEL
MLBr_01159|M.leprae_Br4923 DIAAAFAAADVQILNQDVTAAFERLIVLVRSTSGDERSSVRTRLIELFEL
MAV_1545|M.avium_104 DIDAAFAAADVQILNQDVGAAFDRLIALVRNTSGDDRARVRTRLVELFDL
Mvan_4303|M.vanbaalenii_PYR-1 DIDAAFAAADVEILQQDVQAAFTRLTALVKRTAGDDRTKVRTRLIELFDL
Mflv_2342|M.gilvum_PYR-GCK DIAAALAAADVEILQQQIEPAFARLTALVRRTAGDDRTTVRTRLIELFDL
MSMEG_4917|M.smegmatis_MC2_155 DIEAAFAAADVEILAQQVSAAFDRLIALIKRTAGDDRTKVRTRLIELFEL
TH_1534|M.thermoresistible__bu DIDAAFAAADVEILQQQVTAAFDRLIDLVKRTSGDDRAKVRSRLVELFEL
MAB_1465|M.abscessus_ATCC_1997 DIDAGLAAADVEVLSQQPDSAFDRLVALVRGTSDDDRAKVRARLLELFEL
*: *.:*****::* *: ** ** *:: *:.::*: **:**:***:*
Mb1359|M.bovis_AF2122/97 FDPADPEVVAGRRNLANALY
Rv1324|M.tuberculosis_H37Rv FDPADPEVVAGRRNLANALY
MMAR_4074|M.marinum_M FDPADPEVIAGRRSLASALY
MUL_3937|M.ulcerans_Agy99 FDPADPEVIAGRRSLASALY
MLBr_01159|M.leprae_Br4923 FDPDDPNVIVGRRNLANALY
MAV_1545|M.avium_104 FDPADPEVVAGRRNLANALY
Mvan_4303|M.vanbaalenii_PYR-1 FDPADPEVVAGRRNLANALY
Mflv_2342|M.gilvum_PYR-GCK FDPADPAVIAGRRNLANALY
MSMEG_4917|M.smegmatis_MC2_155 FDPADPEVIAGRRNLANALY
TH_1534|M.thermoresistible__bu FDPSDPQVLTARRNLASALY
MAB_1465|M.abscessus_ATCC_1997 FDPADPAVIAGRRKLANALF
*** ** *:..**.**.**: