For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VRPRRLLIRYAAGLTSAYVLTLAEVVAIVVSLAGRSVVTVGNVITLTVVGLVGTATVALGAVLIVRPSLR WLDSGRPPTPAEIRTTAKTMRRQAAITLAPWLLTAAILVPLNLHAPATVFVVLTTALLFGAIATVSTGYL FTLRTVRPLLAGVPADAAPPGSPGVRVRLVLTWVVCTALPGSAIAVLLILRSRNWLLDESSPIELALVVL ALVAVVLGLRSMLLVSMSISDPLDEVADAMADVEKGKLDRTIDVYEWSEIGRLQSGFNRMVAGLRERDRL RDLFGRHVGEEVARRAFEQATAQNDASTGDERDVAILFIDLVGSTQLANEREPHEVAEVLNDFFEIVVAA VDESHGLVNKFQGDAALAVFGAPLRIDDPASAALSTARILARQLHKLPVDFGIGVSAGPVFAGNIGARNR YEYTVVGDAVNEAARLADRAKEFDGRTLCSGTVLSQASGAEQRLWAAHGEARLRGRSESTDLYTPSGS
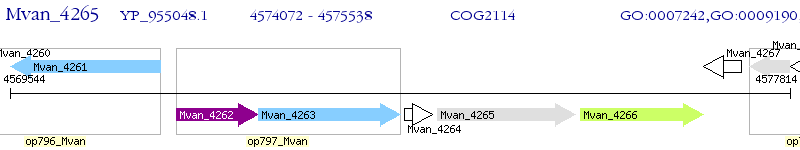
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_4265 | - | - | 100% (488) | putative adenylate/guanylate cyclase |
| M. vanbaalenii PYR-1 | Mvan_5395 | - | 1e-83 | 42.56% (430) | putative adenylate/guanylate cyclase |
| M. vanbaalenii PYR-1 | Mvan_4309 | - | 5e-79 | 39.10% (491) | putative adenylate/guanylate cyclase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3669 | - | 2e-83 | 42.18% (467) | transmembrane protein |
| M. gilvum PYR-GCK | Mflv_2382 | - | 0.0 | 79.34% (484) | putative adenylate/guanylate cyclase |
| M. tuberculosis H37Rv | Rv3645 | - | 2e-83 | 42.18% (467) | transmembrane protein |
| M. leprae Br4923 | MLBr_00201 | - | 3e-84 | 42.58% (458) | hypothetical protein MLBr_00201 |
| M. abscessus ATCC 19977 | MAB_0490c | - | 2e-85 | 41.47% (463) | putative adenylate cyclase |
| M. marinum M | MMAR_4370 | - | 1e-128 | 51.33% (489) | hypothetical protein MMAR_4370 |
| M. avium 104 | MAV_1220 | - | 1e-131 | 51.86% (484) | adenylate cyclase, family protein 3 |
| M. smegmatis MC2 155 | MSMEG_6154 | - | 3e-82 | 41.18% (442) | adenylate cyclase, family protein 3 |
| M. thermoresistible (build 8) | TH_3295 | - | 1e-126 | 49.70% (495) | PUTATIVE putative putative adenylate/guanylate cyclase |
| M. ulcerans Agy99 | MUL_4221 | - | 7e-85 | 43.45% (458) | adenylate cyclase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3669|M.bovis_AF2122/97 MDAEAFVGFRQVPAARYGGLMATTAA-LPRRIHAFVRWVVRTPWPLFSLS
Rv3645|M.tuberculosis_H37Rv MDAEAFVGFRQVPAARYGGLMATTAA-LPRRIHAFVRWVVRTPWPLFSLS
MLBr_00201|M.leprae_Br4923 --------------------MATQAAGLPGRIGAFVRWGIRTPWPLFSLS
MUL_4221|M.ulcerans_Agy99 --------------------MATVAT-LPGRINGFARWVVRTPWPVFSLS
MSMEG_6154|M.smegmatis_MC2_155 ---------------------MTTEPIPIARIGAFARWVARTPWPVFTLG
MAB_0490c|M.abscessus_ATCC_199 ----------------MTGAEGRPDRRPLARAARYSRWVMRTPWPLFIMG
Mvan_4265|M.vanbaalenii_PYR-1 -------------------------------------MRPRRLLIRYAAG
Mflv_2382|M.gilvum_PYR-GCK -------------------------------------MRPRRLLTRYAAG
MMAR_4370|M.marinum_M ------------------------MPSTSASAGQAQPKTARRLLLQYAFD
MAV_1220|M.avium_104 --------------------------------------------------
TH_3295|M.thermoresistible__bu --------------------------VTGARRARADQVGANRIGVTYAAR
Mb3669|M.bovis_AF2122/97 MLQSDIIGALFVLGFLRYGLPPQDNIQLQDLPPVNLLIFVSTVIILFLAG
Rv3645|M.tuberculosis_H37Rv MLQSDIIGALFVLGFLRYGLPPQDNIQLQDLPPVNLLIFVSTVIILFLAG
MLBr_00201|M.leprae_Br4923 MLQSDIIGALFVLGFLRYGLPPQDRVELQDLPIRNLAISTLSVVVAFSTG
MUL_4221|M.ulcerans_Agy99 MLQADIIGGLFVLGFLRYGLPPNDRIQLQDLPPLNLAIFLTSVITLFLAG
MSMEG_6154|M.smegmatis_MC2_155 MLQADIIGALFVLGFLRFGLPPEDRIQLQDLPAFNLAIFLAYLFVSFTVG
MAB_0490c|M.abscessus_ATCC_199 IAQSNLVGALFVLGFLRFGLPPSDRVELTELGPTRLIILGFSLFILFVVG
Mvan_4265|M.vanbaalenii_PYR-1 LTSAYVLTLAEVVAIVVSLAGRSVV------TVGNVITLTVVGLVGTATV
Mflv_2382|M.gilvum_PYR-GCK LTSAYLLTVAEVVAIVVALAGRSVV------TLGNVLTLAVVGLIGTVTV
MMAR_4370|M.marinum_M LALAYFIAVVDASAILIPLRGHTYV----DFAQRNLPVVSVLVVLGIISV
MAV_1220|M.avium_104 MSLSYVLAVGEAAAILIPLRGHTSVGVNADFARQNTGPVLALIALGIVGV
TH_3295|M.thermoresistible__bu LTAAHLTAAAAVLAVIVVLADNTVGDVRRLLTRENLIAFVVLVVLSAAVD
: : . . ..: .
Mb3669|M.bovis_AF2122/97 AVVNLKLLMPVFRWQRRDNLLTEPDPAATELARSRALRMPLYRTLISLAV
Rv3645|M.tuberculosis_H37Rv AVVNLKLLMPVFRWQRRDNLLTEPDPAATELARSRALRMPLYRTLISLAV
MLBr_00201|M.leprae_Br4923 FAVNLKLLMPVFRWQRRDNRLAEADPAATELARSRALRMPFYHTIISLVT
MUL_4221|M.ulcerans_Agy99 FALNLKLLVPVFRWQRRDNLVAAADPSATELARSRVLRMPFYRTLISVGS
MSMEG_6154|M.smegmatis_MC2_155 AYLALRLLIPVIRWQRRETVLSEGDPGITELARARALKMPFYRTLISVTN
MAB_0490c|M.abscessus_ATCC_199 AIVSTYLLLPVLRWQYRS---PDFESEGSEAVRVRALQMPFYRTVISVAT
Mvan_4265|M.vanbaalenii_PYR-1 ALGAVLIVRPSLRWLDSG-------RPPTPAEIRTTAKTMRRQAAITLAP
Mflv_2382|M.gilvum_PYR-GCK AVGAVLIVRPSLRWLNAG-------RRPTPDEIRVTAKTMRRQAVITLAP
MMAR_4370|M.marinum_M AVAGALILAPTLQWFVPG-------HPPTKAQREAALRLGGRQSLILVGA
MAV_1220|M.avium_104 AVAGALSLAPTLRWYVAG-------EEPTPQQRDAVMKLAGRQSAILVTA
TH_3295|M.thermoresistible__bu SAVGILTMRPTFRWFAGG-------AGPTPEQQRAALRIPLRQTGIEFAV
: * ::* : . : :: * .
Mb3669|M.bovis_AF2122/97 WATGGGVFILASWSVAKHAAPVVAVATALGATATAIIGYLQSERVLRPVA
Rv3645|M.tuberculosis_H37Rv WATGGGVFILASWSVAKHAAPVVAVATALGATATAIIGYLQSERVLRPVA
MLBr_00201|M.leprae_Br4923 WGIGGAVFIIASWSAARHSAPIVALATALGATATAIIGYLQSERVLRPVA
MUL_4221|M.ulcerans_Agy99 WAIGSVIFIVASWSVARYAAPVVAIATALGAIATAIIGYLQSERVLRPVA
MSMEG_6154|M.smegmatis_MC2_155 WCLGSIVFIVASWPVASHSAPVVAVATALGATATAIIGYLQSERVLRPVA
MAB_0490c|M.abscessus_ATCC_199 WLVGGAIFVAVTWSSTHSSVLVAAFATVLGATTTSIIGYLQSERVLRPVA
Mvan_4265|M.vanbaalenii_PYR-1 WLLTAAILVPLNLHAPATVFVVLTTALLFGAIATVSTGYLFTLRTVRPLL
Mflv_2382|M.gilvum_PYR-GCK WLLTAAILVPLNLHAPATVFVVLTTALLFGAIATVTTGFLFTLRTLRPIM
MMAR_4370|M.marinum_M WAASGAVLMLLNHADGTKILLPILLGVVLGGPAAAGTGMLLAQRTLRPIV
MAV_1220|M.avium_104 WAVSGGIFLLLNLAGGARLLLPIALGALLGGSAAAGTGMLLAQRTLRPIM
TH_3295|M.thermoresistible__bu WIVSGTVFMVLNLQAPGGVAFVIGGAILFGAMTTACIGYLITTRTLRPII
* . ::: . . :*. :: * * : *.:**:
Mb3669|M.bovis_AF2122/97 VAALRSGVPENVNAPGVILRLMLAWIPSTGVPLLAIVLAVAADKIALLHA
Rv3645|M.tuberculosis_H37Rv VAALRSGVPENVNAPGVILRLMLAWIPSTGVPLLAIVLAVAADKIALLHA
MLBr_00201|M.leprae_Br4923 VAALRSGVPENVKTPGVTLRLMLTWVPSTGVPVLAIVLAVVADKIALLHA
MUL_4221|M.ulcerans_Agy99 VAALRSGVPENLRSPGVISRLMLAWVLSTAVPLLAIVLAVVADKMAILHA
MSMEG_6154|M.smegmatis_MC2_155 VAALRGGVPENFRAPGVILRQVLTWVLSTGVPLLAIILALVASKFSILTA
MAB_0490c|M.abscessus_ATCC_199 IQALRRGVPENVTAPGVIVRQILTWALSTGVPILAIVLTVVGQRAGYLKS
Mvan_4265|M.vanbaalenii_PYR-1 AGVPADAAP--PGSPGVRVRLVLTWVVCTALPGSAIAVLLILRSRNWLLD
Mflv_2382|M.gilvum_PYR-GCK AGVPTDAER--PVAPGVRARLLLMWLACTALPGSAIAVLLILRSQNWLLK
MMAR_4370|M.marinum_M GVATRGGQPR-LAVPGVFARLVLLWFLCSALPIGVIGTFIVLRSYGWVIE
MAV_1220|M.avium_104 RAATLGAEPR-LAVPSVYARLVLLWFLCSAFPIAVIAALVVLRSYGWLVE
TH_3295|M.thermoresistible__bu AAAMTNSTAR-IQLPGPLGRLLYVWILFSVLPALGVVTIVLARSRGWFLQ
. . *. * : * : .* : : .
Mb3669|M.bovis_AF2122/97 TPEALFNPILMMALAALGIGSVSTLLVAMSIADPLRQLRWALSEVQRGNY
Rv3645|M.tuberculosis_H37Rv TPEALFNPILMMALAALGIGSVSTLLVAMSIADPLRQLRWALSEVQRGNY
MLBr_00201|M.leprae_Br4923 VPEQLFSPILLLALAVLVVGLVSTWLVAMSIADPLRQLRWALSEVQRGNY
MUL_4221|M.ulcerans_Agy99 TPDKLFNPILLLALAALAIGSASTLLVSMSIADPLRQLRWALSEVQRGNY
MSMEG_6154|M.smegmatis_MC2_155 SADRLNTPILLLAVAALVIGMSSTVLVAMSIADPLRQLRWALGEVQRGNY
MAB_0490c|M.abscessus_ATCC_199 NADNLTGSILILAIIALAIGLMGTLLVATSIADPLRQLRWALSEVQRGNY
Mvan_4265|M.vanbaalenii_PYR-1 ESSPIELALVVLALVAVVLGLRSMLLVSMSISDPLDEVADAMADVEKGKL
Mflv_2382|M.gilvum_PYR-GCK PTSPIELALLILALVAVVLGLRSMILVSMSISDPVNEVVEAMADVERGEI
MMAR_4370|M.marinum_M QSASLDVPILVVSLAALLLGLPTMILTSRSISDPLDEIVDAMGQVEHGHI
MAV_1220|M.avium_104 KSASLDVPILVVSLAALVLGLPTMILTSRSISDPIGEVVDAMAEIEHGRM
TH_3295|M.thermoresistible__bu ASAPIERPILVMMGVSVVLGISAAVLVARSISDPVHEVVLAMREVERGRP
. : .:::: : :* *.: **:**: :: *: ::::*.
Mb3669|M.bovis_AF2122/97 NAHMQIYDASELGLLQAGFNDMVRELSERQRLRDLFGRYVGEDVARRALE
Rv3645|M.tuberculosis_H37Rv NAHMQIYDASELGLLQAGFNDMVRELSERQRLRDLFGRYVGEDVARRALE
MLBr_00201|M.leprae_Br4923 NAHMQIYDASELGLLQAGFNDMVRELSERQRLRDLFGRYVGEDVARRALE
MUL_4221|M.ulcerans_Agy99 NAHMQIYDASELGLLQAGFNDMVRDLSERQRLRDLFGRYVGEDVARRAIE
MSMEG_6154|M.smegmatis_MC2_155 NAHMQIYDASELGLLQAGFNDMVRDLAERQRLRDLFGRYVGEDVARRALE
MAB_0490c|M.abscessus_ATCC_199 NAHVPIYDASELGLLQAGFNDMVRDLGERQRLRDLFGRYVGEDVARRALE
Mvan_4265|M.vanbaalenii_PYR-1 DRTIDVYEWSEIGRLQSGFNRMVAGLRERDRLRDLFGRHVGEEVARRAFE
Mflv_2382|M.gilvum_PYR-GCK DRTIEVYEWSEIGRLQSGFNRMVAGLRERDRLRDLFGRHVGEEVARRAVD
MMAR_4370|M.marinum_M GTYVGSYERSQIGRLQTGFNRMVAGLEERERLRDLFGRHVGTDVARRALE
MAV_1220|M.avium_104 ETYVGAYERSQIGRLQTGFNRMVAGLAERDRLRDLFGRHVGADVAQRAIE
TH_3295|M.thermoresistible__bu -AAVDVYEPSEIGRLQAGFNSMVAGLAERERLRDLFGRYVGTDVAREAVE
: *: *::* **:*** ** * **:********:** :**:.*.:
Mb3669|M.bovis_AF2122/97 RGTE----LGGQERDVAVLFVDLVGSTQLAATRPPAEVVQLLNEFFRVVV
Rv3645|M.tuberculosis_H37Rv RGTE----LGGQERDVAVLFVDLVGSTQLAATRPPAEVVQLLNEFFRVVV
MLBr_00201|M.leprae_Br4923 HGTE----LGGQERDVAVLFVDLIGSTQLAETKPPTKVVHLLNEFFRVVV
MUL_4221|M.ulcerans_Agy99 RGTE----LGGQERDVAVLFVDLVGSTQLAATRPPVEVVGLLNEFFRVVV
MSMEG_6154|M.smegmatis_MC2_155 RGTE----LGGQERDVAVLFVDLVGSTHLASTIPAADVVNLLNEFFRVIV
MAB_0490c|M.abscessus_ATCC_199 YGTE----LGGQERSVAVLFVDLVGSTQLASTRGPAVVVRVLNEFFRVIV
Mvan_4265|M.vanbaalenii_PYR-1 QATAQNDASTGDERDVAILFIDLVGSTQLANEREPHEVAEVLNDFFEIVV
Mflv_2382|M.gilvum_PYR-GCK LAGEQDDLAAGDEREVAVLFIDLVGSTQLAVEREPRDVAAVLNEFFQIVV
MMAR_4370|M.marinum_M EGSG----SPGEVVEAAVLYIDLVGSTQLAENRPPQEVAEVLNDFFRIVV
MAV_1220|M.avium_104 EGAS----LSGDVVEAAVLFIDLVGSTQLAESRPPQEVAEVLNDFFRIVV
TH_3295|M.thermoresistible__bu HGEA----LRGDVREVGVLFVDLVGSTTLAASRPPEDVARILNDFFQIVV
. *: ...:*::**:*** ** . *. :**:**.::*
Mb3669|M.bovis_AF2122/97 ETVARHGGFVNKFQGDAALAIFGAPIEHPDGAGAALSAARELHDELIPVL
Rv3645|M.tuberculosis_H37Rv ETVARHGGFVNKFQGDAALAIFGAPIEHPDGAGAALSAARELHDELIPVL
MLBr_00201|M.leprae_Br4923 DTVGRHGGFVNKFQGDAALAIFGAPIEHPDSAGAALSAARELHDELLPVL
MUL_4221|M.ulcerans_Agy99 DTVARHGGFVNKFQGDAALAIFGAPIEHPDGAGAALRAARELHDELIPVL
MSMEG_6154|M.smegmatis_MC2_155 DTVNKHGGFVNKFQGDAALAIFGAPIEHPDASGGALAASRELHDELIKVL
MAB_0490c|M.abscessus_ATCC_199 DTVNKHGGFVNKFQGDAALAIFGAPIDHPDGAAAALAAARELSGELTNVL
Mvan_4265|M.vanbaalenii_PYR-1 AAVDESHGLVNKFQGDAALAVFGAPLRIDDPASAALSTARILARQLHKLP
Mflv_2382|M.gilvum_PYR-GCK SEVDKRHGLINKFQGDAALAVFGAPLRIPDPASAAMATARVLASELRRLP
MMAR_4370|M.marinum_M DAVDEHQGLINKFEGDAALAVFGAPLRTGDPASAALATARALGTQLRGLP
MAV_1220|M.avium_104 NAVDEHHGLVNKFAGDAALAVFGVPLPTNEPASAALATARTLGTQLRQLP
TH_3295|M.thermoresistible__bu TAVEDGAGLVNKFQGDAVLAVFGAPLRIDDPAAAALRTARRLASRLRDLP
* *::*** ***.**:**.*: : :..*: ::* * .* :
Mb3669|M.bovis_AF2122/97 G-SAEFGIGVSAGRAIAGHIGAQARFEYTVIGDPVNEAARLTELAKLEDG
Rv3645|M.tuberculosis_H37Rv G-SAEFGIGVSAGRAIAGHIGAQARFEYTVIGDPVNEAARLTELAKLEDG
MLBr_00201|M.leprae_Br4923 G-SAEFGIGVSAGRAIAGHIGAQARFEYTVIGDPVNEAARLTELAKLEDG
MUL_4221|M.ulcerans_Agy99 G-SAEFGIGVSAGRAIAGHIGAQARFEYTVIGDPVNEAARLTELAKLEEG
MSMEG_6154|M.smegmatis_MC2_155 GTETEFGIGVSAGRAIAGHIGAKARFEYTVIGDPVNEAARLTELAKLEEG
MAB_0490c|M.abscessus_ATCC_199 G-SNDFGIGVSAGRAIAGHIGAQARFEYTVIGDPVNEAARLTELAKNEPG
Mvan_4265|M.vanbaalenii_PYR-1 ---VDFGIGVSAGPVFAGNIGARNRYEYTVVGDAVNEAARLADRAKEFDG
Mflv_2382|M.gilvum_PYR-GCK ---VDFGIGISAGPVFAGNIGGRDRYEYTVVGDAVNEAARLADHAKDFDG
MMAR_4370|M.marinum_M A--VDFGVGVSAGRVFAGNIGAENRYEYTVIGDAVNEAARLADLAKTSER
MAV_1220|M.avium_104 ---VDFGIGVSAGRVFAGNVGAENRYEYTVIGDAVNEAARLADLAKTADR
TH_3295|M.thermoresistible__bu G--LDFGIGVSCGRVFAGTIGGEHRYEYTVIGDPVNEASRLADRAKHVAT
:**:*:*.* .:** :*.. *:****:**.****:**:: **
Mb3669|M.bovis_AF2122/97 HVLASAIAVSGALDAEALCWDVGEVVELRGRAAPTQLARPMNLAAPEEVS
Rv3645|M.tuberculosis_H37Rv HVLASAIAVSGALDAEALCWDVGEVVELRGRAAPTQLARPMNLAAPEEVS
MLBr_00201|M.leprae_Br4923 HVLASAIAVSGALDAEALCWNVGEIVELRGRTAPTQLARPLNLAVPEQIT
MUL_4221|M.ulcerans_Agy99 HVLASAFAISGALDAEALCWDVAEVVELRGRTAPTQLARPLNLAVAQKVS
MSMEG_6154|M.smegmatis_MC2_155 HVLASAIAVSGALDSEALCWDVGEIVELRGRTVPTQLARPINLARPEDLD
MAB_0490c|M.abscessus_ATCC_199 NVLASASAVMEARDEEALRWDVGETVELRGRTVSTQVARPRHG-------
Mvan_4265|M.vanbaalenii_PYR-1 RTLCSGTVLSQASGAEQRLWAAHGEARLRGRSESTDLYTPSGS-------
Mflv_2382|M.gilvum_PYR-GCK RTLCSGTAHRLADSRERAYWSADGRTVLRGRSEVTDIFSLTKR-------
MMAR_4370|M.marinum_M RILCSAAALDRAHEAECRRWIQCYSTVLRGRSEATHVCAPAEAG------
MAV_1220|M.avium_104 RILCSAAAIEAAGEAERGHWAECYSTVLRGRSETTHVSAPTG--------
TH_3295|M.thermoresistible__bu RVLASGAALAAAQPEEHTRWVGRGSALLRGRAEPTELAEPAGPDDPAAG-
. *.*. . * * * . ****: *.:
Mb3669|M.bovis_AF2122/97 SEVRG---------------------------------
Rv3645|M.tuberculosis_H37Rv SEVRG---------------------------------
MLBr_00201|M.leprae_Br4923 SEVTG---------------------------------
MUL_4221|M.ulcerans_Agy99 TKSGS---------------------------------
MSMEG_6154|M.smegmatis_MC2_155 GSANGQSTDGSPAETAFQQEKGLFAVEGVEVVGASVPG
MAB_0490c|M.abscessus_ATCC_199 --------------------------------------
Mvan_4265|M.vanbaalenii_PYR-1 --------------------------------------
Mflv_2382|M.gilvum_PYR-GCK --------------------------------------
MMAR_4370|M.marinum_M --------------------------------------
MAV_1220|M.avium_104 --------------------------------------
TH_3295|M.thermoresistible__bu --------------------------------------