For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
GVRVLVAKPGLDGHDRGAKIVARTLRDAGFEVIYTGIRQRIEDIVSIALQEDVALVGLSILSGAHVALTT RTVDALRAADAGDIAVVVGGTIPQGDVQKLLDAGAAAVFPTGTSLEDLVRDVRALTEKVEQS*
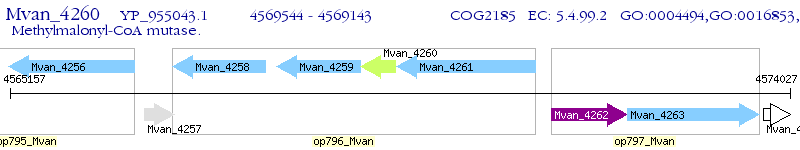
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_4260 | - | - | 100% (133) | cobalamin B12-binding domain-containing protein |
| M. vanbaalenii PYR-1 | Mvan_2763 | - | 2e-22 | 44.35% (115) | methylmalonyl-CoA mutase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1530 | mutB | 1e-22 | 41.67% (120) | methylmalonyl-CoA mutase |
| M. gilvum PYR-GCK | Mflv_2388 | - | 3e-65 | 94.74% (133) | cobalamin B12-binding domain-containing protein |
| M. tuberculosis H37Rv | Rv1493 | mutB | 1e-22 | 41.67% (120) | methylmalonyl-CoA mutase |
| M. leprae Br4923 | MLBr_01799 | mutB | 2e-21 | 44.14% (111) | methylmalonyl-CoA mutase |
| M. abscessus ATCC 19977 | MAB_2711c | - | 1e-22 | 45.22% (115) | methylmalonyl-CoA mutase |
| M. marinum M | MMAR_4797 | mcmA2b | 3e-56 | 85.04% (127) | methylmalonyl-CoA mutase alpha subunit, McmA2b |
| M. avium 104 | MAV_0857 | - | 2e-55 | 84.25% (127) | putative methylmalonyl-CoA mutase |
| M. smegmatis MC2 155 | MSMEG_4880 | - | 2e-61 | 92.31% (130) | methylmalonyl-CoA mutase C-terminal domain-containing protein |
| M. thermoresistible (build 8) | TH_0128 | - | 9e-56 | 88.00% (125) | methylmalonyl-CoA mutase C-terminal domain protein |
| M. ulcerans Agy99 | MUL_0366 | mcmA2b | 3e-55 | 83.46% (127) | methylmalonyl-CoA mutase alpha subunit, McmA2b |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_4260|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2388|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_4880|M.smegmatis_MC2_155 --------------------------------------------------
TH_0128|M.thermoresistible__bu --------------------------------------------------
MMAR_4797|M.marinum_M --------------------------------------------------
MUL_0366|M.ulcerans_Agy99 --------------------------------------------------
MAV_0857|M.avium_104 --------------------------------------------------
Mb1530|M.bovis_AF2122/97 MTTKTP----------VIGSFAGVPLHSERAAQSPTEAAVHTHVAAAAAA
Rv1493|M.tuberculosis_H37Rv MTTKTP----------VIGSFAGVPLHSERAAQSPTEAAVHTHVAAAAAA
MLBr_01799|M.leprae_Br4923 MTVRDVGGQQDRETTFVVGSFADIPLRSDRVVWPPSEAAVEKHVFSAASA
MAB_2711c|M.abscessus_ATCC_199 MTDLTDLS----ESGTILGSFADVPLHGDRVSVAPDTAAVQTAVSEAAAA
Mvan_4260|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2388|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_4880|M.smegmatis_MC2_155 --------------------------------------------------
TH_0128|M.thermoresistible__bu --------------------------------------------------
MMAR_4797|M.marinum_M --------------------------------------------------
MUL_0366|M.ulcerans_Agy99 --------------------------------------------------
MAV_0857|M.avium_104 --------------------------------------------------
Mb1530|M.bovis_AF2122/97 HGYTPEQLVWHTPEGIDVTPVYIAADRAAAEAEGYPLHSFPGEPPFVRGP
Rv1493|M.tuberculosis_H37Rv HGYTPEQLVWHTPEGIDVTPVYIAADRAAAEAEGYPLHSFPGEPPFVRGP
MLBr_01799|M.leprae_Br4923 HGYTVDQLHWHTPEGIDVKPVYIAADRAAAEAGGYPLHSFPGEPPYLRGP
MAB_2711c|M.abscessus_ATCC_199 HGYSPEQLDWETPEAITVKPIYTAADREAVRAQGYPLDTFPGQVPFLRGP
Mvan_4260|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2388|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_4880|M.smegmatis_MC2_155 --------------------------------------------------
TH_0128|M.thermoresistible__bu --------------------------------------------------
MMAR_4797|M.marinum_M --------------------------------------------------
MUL_0366|M.ulcerans_Agy99 --------------------------------------------------
MAV_0857|M.avium_104 --------------------------------------------------
Mb1530|M.bovis_AF2122/97 YPTMYVNQPWTIRQYAGFSTAADSNAFYRRNLAAGQKGLSVAFDLATHRG
Rv1493|M.tuberculosis_H37Rv YPTMYVNQPWTIRQYAGFSTAADSNAFYRRNLAAGQKGLSVAFDLATHRG
MLBr_01799|M.leprae_Br4923 YPTMYVNQPWTIRQYAGFSTAADSNAFYRRNLAAGQKGLSVAFDLATHRG
MAB_2711c|M.abscessus_ATCC_199 YPTMYVNQPWTIRQYAGFSTAAESNAFYRRNLAAGQKGLSVAFDLATHRG
Mvan_4260|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2388|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_4880|M.smegmatis_MC2_155 --------------------------------------------------
TH_0128|M.thermoresistible__bu --------------------------------------------------
MMAR_4797|M.marinum_M --------------------------------------------------
MUL_0366|M.ulcerans_Agy99 --------------------------------------------------
MAV_0857|M.avium_104 --------------------------------------------------
Mb1530|M.bovis_AF2122/97 YDSDHPRVQGDVGMAGVAIDSILDMRQLFDGIDLSTVSVSMTMNGAVLPI
Rv1493|M.tuberculosis_H37Rv YDSDHPRVQGDVGMAGVAIDSILDMRQLFDGIDLSTVSVSMTMNGAVLPI
MLBr_01799|M.leprae_Br4923 YDSDHPRVQGDVGMAGVAIDSILDMRQLFDGIDLSAVSVSMTMNGAVLPI
MAB_2711c|M.abscessus_ATCC_199 YDSDHPRVQGDVGMAGVAIDSILDMRQLFDGIDLGSVSVSMTMNGAVLPI
Mvan_4260|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2388|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_4880|M.smegmatis_MC2_155 --------------------------------------------------
TH_0128|M.thermoresistible__bu --------------------------------------------------
MMAR_4797|M.marinum_M --------------------------------------------------
MUL_0366|M.ulcerans_Agy99 --------------------------------------------------
MAV_0857|M.avium_104 --------------------------------------------------
Mb1530|M.bovis_AF2122/97 LALYVVAAEEQGVAPEQLAGTIQNDILKEFMVRNTYIYPPKPSMRIISDI
Rv1493|M.tuberculosis_H37Rv LALYVVAAEEQGVAPEQLAGTIQNDILKEFMVRNTYIYPPKPSMRIISDI
MLBr_01799|M.leprae_Br4923 LALYVVAAEEQGVPPEKLAGTIQNDILKEFMVRNTYIYPPEPSMRIISDI
MAB_2711c|M.abscessus_ATCC_199 LALYVVAAEEQGVGTEKLAGTIQNDILKEFMVRNTYIYPPKPSMRIISDI
Mvan_4260|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2388|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_4880|M.smegmatis_MC2_155 --------------------------------------------------
TH_0128|M.thermoresistible__bu --------------------------------------------------
MMAR_4797|M.marinum_M --------------------------------------------------
MUL_0366|M.ulcerans_Agy99 --------------------------------------------------
MAV_0857|M.avium_104 --------------------------------------------------
Mb1530|M.bovis_AF2122/97 FAYTSAKMPKFNSISISGYHIQEAGATADLELAYTLADGVDYIRAGLNAG
Rv1493|M.tuberculosis_H37Rv FAYTSAKMPKFNSISISGYHIQEAGATADLELAYTLADGVDYIRAGLNAG
MLBr_01799|M.leprae_Br4923 FAYTSAKMPKFNSISISGYHIQEAGATADLELAYTLADGVDYIKAGLDAG
MAB_2711c|M.abscessus_ATCC_199 FAYTSAKMPKFNSISISGYHIQEAGATADLELAYTLADGVEYIKAGLAGG
Mvan_4260|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2388|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_4880|M.smegmatis_MC2_155 --------------------------------------------------
TH_0128|M.thermoresistible__bu --------------------------------------------------
MMAR_4797|M.marinum_M --------------------------------------------------
MUL_0366|M.ulcerans_Agy99 --------------------------------------------------
MAV_0857|M.avium_104 --------------------------------------------------
Mb1530|M.bovis_AF2122/97 LDIDSFAPRLSFFWGIGMNFFMEVAKLRAGRLLWSELVAQFAPKSAKSLS
Rv1493|M.tuberculosis_H37Rv LDIDSFAPRLSFFWGIGMNFFMEVAKLRAGRLLWSELVAQFAPKSAKSLS
MLBr_01799|M.leprae_Br4923 LDIDTFAPRLSFFWGIGMNFFMEVAKLRAGRLLWSELVAKFGPQSPKSLS
MAB_2711c|M.abscessus_ATCC_199 LDIDVFAPRLSFFWAIGMNFFMEVAKLRAGRLLWSELVAQFDPKSSKSQS
Mvan_4260|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2388|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_4880|M.smegmatis_MC2_155 --------------------------------------------------
TH_0128|M.thermoresistible__bu --------------------------------------------------
MMAR_4797|M.marinum_M --------------------------------------------------
MUL_0366|M.ulcerans_Agy99 --------------------------------------------------
MAV_0857|M.avium_104 --------------------------------------------------
Mb1530|M.bovis_AF2122/97 LRTHSQTSGWSLTAQDVFNNVARTCIEAMAATQGHTQSLHTNALDEALAL
Rv1493|M.tuberculosis_H37Rv LRTHSQTSGWSLTAQDVFNNVARTCIEAMAATQGHTQSLHTNALDEALAL
MLBr_01799|M.leprae_Br4923 LRTHSQTSGWSLTAQDPFNNVARTCIEAMAATQGHTQSLHTNALDEALAL
MAB_2711c|M.abscessus_ATCC_199 LRTHSQTSGWSLTAQDVFNNVARTCIEAMAATQGHTQSLHTNALDEALAL
Mvan_4260|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2388|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_4880|M.smegmatis_MC2_155 --------------------------------------------------
TH_0128|M.thermoresistible__bu --------------------------------------------------
MMAR_4797|M.marinum_M --------------------------------------------------
MUL_0366|M.ulcerans_Agy99 --------------------------------------------------
MAV_0857|M.avium_104 --------------------------------------------------
Mb1530|M.bovis_AF2122/97 PTDFSARIARNTQLVLQQESGTTRPIDPWGGSYYVEWLTHRLARRARAHI
Rv1493|M.tuberculosis_H37Rv PTDFSARIARNTQLVLQQESGTTRPIDPWGGSYYVEWLTHRLARRARAHI
MLBr_01799|M.leprae_Br4923 PTDFSARIARNTQLVLQQESGTTRPIDPWGGSYYVEWLTHQLAARARTHI
MAB_2711c|M.abscessus_ATCC_199 PTDFSARIARNTQLLIQQESGTCRPIDPWGGSYYVEWLTHQLASRAREHI
Mvan_4260|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2388|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_4880|M.smegmatis_MC2_155 --------------------------------------------------
TH_0128|M.thermoresistible__bu --------------------------------------------------
MMAR_4797|M.marinum_M --------------------------------------------------
MUL_0366|M.ulcerans_Agy99 --------------------------------------------------
MAV_0857|M.avium_104 --------------------------------------------------
Mb1530|M.bovis_AF2122/97 AEVAEHGGMAQAISDGIPKLRIEEAAARTQARIDSGQQPVVGVNKYQVPE
Rv1493|M.tuberculosis_H37Rv AEVAEHGGMAQAISDGIPKLRIEEAAARTQARIDSGQQPVVGVNKYQVPE
MLBr_01799|M.leprae_Br4923 AEVAEHGGMAAAISDGIPKLRIEEAAARTQARIDSGRQPVVGVNKYQVDQ
MAB_2711c|M.abscessus_ATCC_199 REVTEAGGMAQAIDQGIPKLRIEEAAARTQARIDSGRQPVIGVNKYQVAE
Mvan_4260|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2388|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_4880|M.smegmatis_MC2_155 --------------------------------------------------
TH_0128|M.thermoresistible__bu --------------------------------------------------
MMAR_4797|M.marinum_M --------------------------------------------------
MUL_0366|M.ulcerans_Agy99 --------------------------------------------------
MAV_0857|M.avium_104 --------------------------------------------------
Mb1530|M.bovis_AF2122/97 DHEIEVLKVENSRVRAEQLAKLQRLRAGRDEPAVRAALAELTRAAAEQGR
Rv1493|M.tuberculosis_H37Rv DHEIEVLKVENSRVRAEQLAKLQRLRAGRDEPAVRAALAELTRAAAEQGR
MLBr_01799|M.leprae_Br4923 DHEIEVLKVENSRVRAEQVAKLQQLRANRDQLAVDAALAELSGAAGAHGR
MAB_2711c|M.abscessus_ATCC_199 DQQIEVLKVENSRVRAEQLAKLQQLRAERDDDAVQAALAELTRAAADSGR
Mvan_4260|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2388|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_4880|M.smegmatis_MC2_155 --------------------------------------------------
TH_0128|M.thermoresistible__bu --------------------------------------------------
MMAR_4797|M.marinum_M --------------------------------------------------
MUL_0366|M.ulcerans_Agy99 --------------------------------------------------
MAV_0857|M.avium_104 --------------------------------------------------
Mb1530|M.bovis_AF2122/97 AGADGLGNNLLALAIDAARAQATVGEISEALEKVYGRHRAEIRTISGVYR
Rv1493|M.tuberculosis_H37Rv AGADGLGNNLLALAIDAARAQATVGEISEALEKVYGRHRAEIRTISGVYR
MLBr_01799|M.leprae_Br4923 AG-----DNLLALAINAARVQATVGEISDALERVYGRHQAKIRTITGVYR
MAB_2711c|M.abscessus_ATCC_199 AGEDGLGNNLMALAINAARHKATVGEISDALEKVYGRHQAEIRTISGVYR
Mvan_4260|M.vanbaalenii_PYR-1 ---------------------------MGVRVLVAKPGLDGHDRGAKIVA
Mflv_2388|M.gilvum_PYR-GCK ---------------------------MSTRVLVAKPGLDGHDRGAKIVA
MSMEG_4880|M.smegmatis_MC2_155 ---------------------------MATRVLVAKPGLDGHDRGAKIVA
TH_0128|M.thermoresistible__bu ----------------------VGAPDSPIRVLVAKPGLDGHDRGAKIVA
MMAR_4797|M.marinum_M ---------------------------MAARILVAKPGLDGHDRGAKIVA
MUL_0366|M.ulcerans_Agy99 ---------------------------MAARILVAKPGLNGHDRGAKIVA
MAV_0857|M.avium_104 ---------------------------MAVRILVAKPGLDGHDRGAKIVA
Mb1530|M.bovis_AF2122/97 ---DEVGKAPNIAAATELVEKFAEADGRRPRILIAKMGQDGHDRGQKVIA
Rv1493|M.tuberculosis_H37Rv ---DEVGKAPNIAAATELVEKFAEADGRRPRILIAKMGQDGHDRGQKVIA
MLBr_01799|M.leprae_Br4923 REVGEAGKATDIANATELVKKFAEADGRRPRILVAKMGQDGHDRGQKVVA
MAB_2711c|M.abscessus_ATCC_199 DEVGRDGAVSSVTQATELVQKFADADGRRPRVLVAKMGQDGHDRGQKVIA
*:*:** * :***** *::*
Mvan_4260|M.vanbaalenii_PYR-1 RTLRDAGFEVIYTGIRQRIEDIVSIALQEDVALVGLSILSGAHVALTTRT
Mflv_2388|M.gilvum_PYR-GCK RTLRDAGFEVIYTGIRQRIEDIVSIALQEDVALVGLSILSGAHVTLTART
MSMEG_4880|M.smegmatis_MC2_155 RTLRDAGFEVIYTGIRQRIEDIVSIALQEDVALVGLSILSGAHVALTART
TH_0128|M.thermoresistible__bu RTLRDAGFEVIYTGIRQRIEDIVSIALQEDVALVGLSILSGAHLALTRRT
MMAR_4797|M.marinum_M RTLRDAGFEVIYTGIRQRIEDIASIAVQEDVAVVGLSILSGAHLALTART
MUL_0366|M.ulcerans_Agy99 RTLRDAGFEVIYNGIRQRIEDIASIAVQEDVAVVGLSILSGAHLALTART
MAV_0857|M.avium_104 RTLRDAGFEVIYTGIRQRIEDIASIAVQEDVAVVGLSILSGAHLALTART
Mb1530|M.bovis_AF2122/97 TAFADIGFDVDVGSLFSTPEEVARQAADNDVHVIGVSSLAAGHLTLVPAL
Rv1493|M.tuberculosis_H37Rv TAFADIGFDVDVGSLFSTPEEVARQAADNDVHVIGVSSLAAGHLTLVPAL
MLBr_01799|M.leprae_Br4923 TAFADLGFDVDVGSLFSTPEEVARQAADNDVHVIGVSSLAAGHLTLVPAL
MAB_2711c|M.abscessus_ATCC_199 TAFADIGFDVDVGPLFQTPEEVARQAADNDVHVVGVSSLAAGHLTLVPAL
:: * **:* : . *::. * ::** ::*:* *:..*::*.
Mvan_4260|M.vanbaalenii_PYR-1 VDALRAADAGDIAVVVGGTIPQGDVQKLLDAGAAAVFPTGTSLEDLVRDV
Mflv_2388|M.gilvum_PYR-GCK VDALRAADAGDIAVVVGGTIPQGDVQKLLDAGAAAVFPTGTSLDDLVRDV
MSMEG_4880|M.smegmatis_MC2_155 VEALRAADAGDIAVVVGGTIPQGDVQKLLDVGAAAVFPTGTPLDTLVTEV
TH_0128|M.thermoresistible__bu VEALRAADAGDIAVVVGGTIPQQDVPKLQEVGAAAVFPTGTPLDVLVTEV
MMAR_4797|M.marinum_M IEALRDADAADIAVVVGGTIPHSDVPKLLAAGAAAVFPTGTPLDDLVRDI
MUL_0366|M.ulcerans_Agy99 IEALRDADAADIAVVVGGTIPHSDVPKLLAAGAAAVFPTGTPLDDLVRDI
MAV_0857|M.avium_104 VEALRACDAADIAVVVGGTIPHADVPKLLAAGAAAVFPTGTPLDKLVREI
Mb1530|M.bovis_AF2122/97 RDALAQVGRPDIMIVVGGVIPPGDFDELYAAGATAIFPPGTVIADAAIDL
Rv1493|M.tuberculosis_H37Rv RDALAQVGRPDIMIVVGGVIPPGDFDELYAAGATAIFPPGTVIADAAIDL
MLBr_01799|M.leprae_Br4923 RDALADVGRPDIMIVVGGVIPPDDFDELYAAGATAIFPPGTVIAYAAIGL
MAB_2711c|M.abscessus_ATCC_199 RQALAEVGRPDIMIVVGGVIPPGDFDELYQAGAAAIFPPGTVISEAAIGL
:** . ** :****.** *. :* .**:*:**.** : . :
Mvan_4260|M.vanbaalenii_PYR-1 R-ALTEKVEQS-------------
Mflv_2388|M.gilvum_PYR-GCK R-ALTEKAEQS-------------
MSMEG_4880|M.smegmatis_MC2_155 R-ALTEKVAK--------------
TH_0128|M.thermoresistible__bu R-KLTGAAADV-------------
MMAR_4797|M.marinum_M R-ALTG-AAPTAPQPSMEEPCVSE
MUL_0366|M.ulcerans_Agy99 R-ALTG-DAQTAPQPSMEEPCVSE
MAV_0857|M.avium_104 R-ALTGTGEPDDKEAITEEQCASE
Mb1530|M.bovis_AF2122/97 LHRLAERLGYTLD-----------
Rv1493|M.tuberculosis_H37Rv LHRLAERLGYTLD-----------
MLBr_01799|M.leprae_Br4923 LHKLAGRLGYTLT-----------
MAB_2711c|M.abscessus_ATCC_199 LHKLAERLGYDLT-----------
*: