For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
THRAVYPRLVRELERPIVVLGRIGDMALFYARAAAGVPYALRFYRRDILRLIAEISMGAGTLAMIGGTLV IVGFLTLATGGTLAVQGYSSLGDIGIEALTGFLAAFINVRISAPVVAGIGLAATFGAGVTAQLGAMRTNE EIDALEAMAIRPISFLVSTRLIAGMVAITPLYSVAVILSFLASRYTTVVLLGQSAGLYDHYFATFLSPID LLWSFAQAVLMAVAILLIHTYFGYNATGGPAGVGVATGDAVRTSLVVVVSVTLIVSLAVYGAGGNFNLAG *
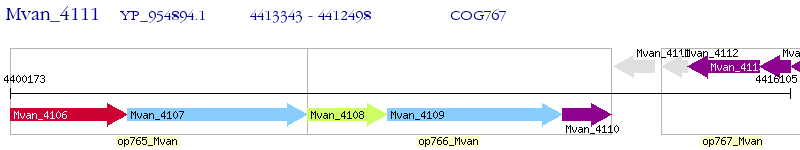
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_4111 | - | - | 100% (281) | hypothetical protein Mvan_4111 |
| M. vanbaalenii PYR-1 | Mvan_2850 | - | e-124 | 78.78% (278) | hypothetical protein Mvan_2850 |
| M. vanbaalenii PYR-1 | Mvan_4151 | - | e-122 | 78.06% (278) | hypothetical protein Mvan_4151 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3530c | yrbE4B | 1e-101 | 64.36% (275) | integral membrane protein YrbE4b |
| M. gilvum PYR-GCK | Mflv_2507 | - | 1e-125 | 79.86% (278) | hypothetical protein Mflv_2507 |
| M. tuberculosis H37Rv | Rv3500c | yrbE4B | 1e-101 | 64.36% (275) | integral membrane protein YrbE4b |
| M. leprae Br4923 | MLBr_02588 | yrbE1B | 4e-80 | 53.57% (280) | hypothetical protein MLBr_02588 |
| M. abscessus ATCC 19977 | MAB_4154c | - | 3e-98 | 66.41% (259) | hypothetical protein MAB_4154c |
| M. marinum M | MMAR_4711 | - | 1e-127 | 80.58% (278) | hypothetical protein MMAR_4711 |
| M. avium 104 | MAV_0947 | - | 1e-123 | 77.34% (278) | TrnB2 protein |
| M. smegmatis MC2 155 | MSMEG_4795 | - | 1e-124 | 78.06% (278) | ABC-transporter integral membrane protein |
| M. thermoresistible (build 8) | TH_0522 | - | 1e-125 | 79.64% (280) | TrnB2 protein |
| M. ulcerans Agy99 | MUL_4063 | yrbE4B | 1e-102 | 64.36% (275) | membrane protein YrbE4B |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3530c|M.bovis_AF2122/97 MS------YDVTIRFRRFFSRLQRP----VDNFG---EQALFYGETMRYV
Rv3500c|M.tuberculosis_H37Rv MS------YDVTIRFRRFFSRLQRP----VDNFG---EQALFYGETMRYV
MUL_4063|M.ulcerans_Agy99 MS------YDLTVRFHRFMSRLARP----VDNLG---EQALFYGETVRYI
MAB_4154c|M.abscessus_ATCC_199 MSGPYLSTHTVTSYTVRYMRGLGRS----FDKFG---EQALFYAQSLSYI
MMAR_4711|M.marinum_M --------MALRAVYPRMTRQLEKP----VGTLSRIGDHTLFYGKAIAGA
MAV_0947|M.avium_104 --------MALRAAYPRLTRQLERP----VALLAGIGDHALFYGKALAGM
MSMEG_4795|M.smegmatis_MC2_155 -------MTILRSTYPRLTRQFNKP----VATLSRVGDHTLFYLRAIAGT
TH_0522|M.thermoresistible__bu MG----TIQTLRATYPRLARQIERP----VSLMGRIGDHTLFYGRALAGA
Mflv_2507|M.gilvum_PYR-GCK MG----TSTVIRSRSPRLVGNLGRP----VAVLGRLGDHVLFYGRAVAGV
Mvan_4111|M.vanbaalenii_PYR-1 --------MTHRAVYPRLVRELERP----IVVLGRIGDMALFYARAAAGV
MLBr_02588|M.leprae_Br4923 MS----TAAVLRARFPRAAANLNRYGGAAARGIDDIGQMSWFGLIALGNI
* : : : : * :
Mb3530c|M.bovis_AF2122/97 PNAITRYRKETVRLVAEMTLGAGALVMIGGTVGVAAFLTLASGGVIAVQG
Rv3500c|M.tuberculosis_H37Rv PNAITRYRKETVRLVAEMTLGAGALVMIGGTVGVAAFLTLASGGVIAVQG
MUL_4063|M.ulcerans_Agy99 PNATTRYRKETIRLVAEMTLGAGALVMIGGTVGVAAFLTLASGGVIAVQG
MAB_4154c|M.abscessus_ATCC_199 PAALTKYRKETIRVLAEITMGTGALVMIGGTVGVAVFLTLASGGVIAVQG
MMAR_4711|M.marinum_M PFAATHYRREIIRLVAEISMGAGTLAMIGGTLVIVGFLTLAAGGTLAVQG
MAV_0947|M.avium_104 PFAATRYTREVVRLVAEISMGAGTLAMIGGTVVIVGFLTLAAGGTLAIQG
MSMEG_4795|M.smegmatis_MC2_155 PHAALHYRKELVRLIAEISMGAGTLAMIGGTVVIVGFLTLATGGVLAIQG
TH_0522|M.thermoresistible__bu PYAAMRFRTEIIRLIAEISMGAGTLAMIGGTVVIVGFLTLAAGGTLAVQG
Mflv_2507|M.gilvum_PYR-GCK PHAAVHFRKEIIRLIAEISMGAGTLAMIGGTVVIVGFLTLAAGGTLAIQG
Mvan_4111|M.vanbaalenii_PYR-1 PYALRFYRRDILRLIAEISMGAGTLAMIGGTLVIVGFLTLATGGTLAVQG
MLBr_02588|M.leprae_Br4923 PNALSRYRKETLRLIAQIGMGTGAMAVVGGTAAIVGFVTLSGSSLVAIQG
* * : : :*::*:: :*:*::.::*** :. *:**: .. :*:**
Mb3530c|M.bovis_AF2122/97 YSSLGDIGIEALTGFLSAFLNVRVVAPVIAGIALAATIGAGATAQLGAMR
Rv3500c|M.tuberculosis_H37Rv YSSLGDIGIEALTGFLSAFLNVRVVAPVIAGIALAATIGAGATAQLGAMR
MUL_4063|M.ulcerans_Agy99 YSSLGDIGIEALTGFLSAFLNVRVVAPVIAGIALAATIGAGATAQLGAMR
MAB_4154c|M.abscessus_ATCC_199 YSSLGNIGIEALTGFLSAFLNVRIVAPVVAGIAMAATIGAGTTAQLGAMR
MMAR_4711|M.marinum_M YSSLGNIGIEALTGFLAAFINVRIAAPIVAGIGLAATFGAGVTAQLGAMR
MAV_0947|M.avium_104 YTSLGNIGIEALTGFLAAFINVRIAAPVVAGIGLAATFGAGVTAQLGAMR
MSMEG_4795|M.smegmatis_MC2_155 YSSLGNIGIEALTGFLSAFINVRIAAPVVAGIGLAATFGAGVTAQLGAMR
TH_0522|M.thermoresistible__bu YSSLGDIGIEALTGFLSAFINVRIAAPVVAGIGLAATFGAGVTAQLGAMR
Mflv_2507|M.gilvum_PYR-GCK YSSLGDIGIEALTGFLAAFINVRIAAPVVAGIGLAATFGAGVTAQLGAMR
Mvan_4111|M.vanbaalenii_PYR-1 YSSLGDIGIEALTGFLAAFINVRISAPVVAGIGLAATFGAGVTAQLGAMR
MLBr_02588|M.leprae_Br4923 FASLGAIGVEAFTGFFAALINVRIAAPVITGIVMAATVGAGATAELGAMR
::*** **:**:***::*::***: **:::** :***.***.**:*****
Mb3530c|M.bovis_AF2122/97 VSEEIDAVECMAVHSVSYLVSTRLIAGLVAIIPLYSLSVLAAFFAARFTT
Rv3500c|M.tuberculosis_H37Rv VSEEIDAVECMAVHSVSYLVSTRLIAGLVAIIPLYSLSVLAAFFAARFTT
MUL_4063|M.ulcerans_Agy99 VSEEIDAVECMAVHSVSYLVSTRLIAGLVAIIPLYSLAVLAAFFAARFTT
MAB_4154c|M.abscessus_ATCC_199 VAEEIDALETMAVHAISYLVSTRLVAGLIAIIPLYSLSVLAAFFASRATT
MMAR_4711|M.marinum_M INEEIDALESMAIRPVSYLVSTRIVAGMMAITPLYSIAVILSFLASQFTT
MAV_0947|M.avium_104 INEEVDALESMAIRPVAYLVSTRILAGMLAITPLYSIAVILSFVASQFTT
MSMEG_4795|M.smegmatis_MC2_155 INEEIDALESMAIRPVEYLVSTRIVAGMIAITPLYAIAVILSFLASQFTT
TH_0522|M.thermoresistible__bu INEEVDALESMGIPPIPYLVSTRIVAGMLAITPLYSIAVILSFVASQFTT
Mflv_2507|M.gilvum_PYR-GCK INEEIDALESMGIRPVEYLVSTRIVAGMVAITPLYSIAVILSFIASQFTT
Mvan_4111|M.vanbaalenii_PYR-1 TNEEIDALEAMAIRPISFLVSTRLIAGMVAITPLYSVAVILSFLASRYTT
MLBr_02588|M.leprae_Br4923 ISEEIDALEVMSIKSISFLVTTRILAGLVVIVPLYAVAMIMAFLSPHIIT
**:**:* *.: .: :**:**::**::.* ***::::: :*.:.: *
Mb3530c|M.bovis_AF2122/97 VFVNGQSAGLYDHYFNTFLIPSDLLWSFMQAIAMSIAVMLVHTYYGYNAS
Rv3500c|M.tuberculosis_H37Rv VFVNGQSAGLYDHYFNTFLIPSDLLWSFMQAIAMSIAVMLVHTYYGYNAS
MUL_4063|M.ulcerans_Agy99 VFINGQSAGLYDHYFNTFLIPSDLLWSFLQAIVMAIAVMLVHTYYGYNAS
MAB_4154c|M.abscessus_ATCC_199 VMINGQSPGVYDHYFNTFLVPTDLLWSFLQAIVMSVVVMLVHTSYGFNAS
MMAR_4711|M.marinum_M VILFGQSGGLYQHYFTTFLNPIDLLWSFLQAVLMAITILLIHTYFGYFAS
MAV_0947|M.avium_104 TFLLGQSQGLYQHYFNTFLNPIDLLWSFLQAILMALTILLIHTYYGYFAS
MSMEG_4795|M.smegmatis_MC2_155 VVLLGQSGGLYNHYFDTFLNPIDLLWSFLQAILMAITILLIHTYFGYFAS
TH_0522|M.thermoresistible__bu VVLFGQSSGLYAHYFDTFLNPIDLLWSFLQAVLMAITILLIHTYFGYFAT
Mflv_2507|M.gilvum_PYR-GCK VVLFGQSGGLYDHYFDTFLNPIDLLWSFLQAVLMAIAILLVHTYFGYFAS
Mvan_4111|M.vanbaalenii_PYR-1 VVLLGQSAGLYDHYFATFLSPIDLLWSFAQAVLMAVAILLIHTYFGYNAT
MLBr_02588|M.leprae_Br4923 TVLYGQSNGTYEHYFRTFLRPDDVFWSFLEAVIITAVVMITHCYYGYTAN
..: *** * * *** *** * *::*** :*: :: .::: * :*: *.
Mb3530c|M.bovis_AF2122/97 GGSVGVGVAVGQAVRTSLIVVVVITLFISLAVYGASGNFNLSG
Rv3500c|M.tuberculosis_H37Rv GGSVGVGVAVGQAVRTSLIVVVVITLFISLAVYGASGNFNLSG
MUL_4063|M.ulcerans_Agy99 GGSVGVGIAVGQAVRTSLIVVVVITLFISLAVYGASGNFNLSG
MAB_4154c|M.abscessus_ATCC_199 GGPVGVGVAVGQAVRTSLIVVVLVTLFISLAVYGGSGNFNLSG
MMAR_4711|M.marinum_M GGPAGVGVATGNAVRTSLIVVVSVTLLVSLSVYGSNGNFHLSG
MAV_0947|M.avium_104 GGPAGVGNATGNAVRTSLIVVVSVTLLVSLSIYGTNGNFNLSG
MSMEG_4795|M.smegmatis_MC2_155 GGPSGVGVAVGNAVRTSLIVVVSVTLLVSLAVYGSNGNFNLSG
TH_0522|M.thermoresistible__bu GGPSGVGVAVGNAVRTSLIVVVSVTLLVSLSVYGSNGNFNLSG
Mflv_2507|M.gilvum_PYR-GCK GGPSGVGAAVGNAVRTSLVVVVSVTLLVSLSIYGSNGNFNLSG
Mvan_4111|M.vanbaalenii_PYR-1 GGPAGVGVATGDAVRTSLVVVVSVTLIVSLAVYGAGGNFNLAG
MLBr_02588|M.leprae_Br4923 GGPVGVGEAVGRSMRFSLVSVQVVVLSATLALYGVDPNFALTV
**. *** *.* ::* **: * :.* :*::** . ** *: