For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSRQDISDVVVIGAGIVGCAIARALSGTGLSVTLLEARGDVGDGTSKANTALLHTGFDATPGTLESRLVA RGYQLLGEYAGQTGIPVERTGAMLVAWTEEEAGALPGLMVKAERNGYRACGIVGADEVYRRVPDLGPGAL AGLDVPGEAIICTWTTNLALATDAVARGARLLRGARVTAVETGPEHTTLRTTSGEVRARWVINAAGLGAD HLDAEFGYRRFTVTPRRGELLVFDKLTRPMVPLIVLAVPSSRGKGVLVSPTIYGNVMVGPTSENLEDRTA TGTSEKGFEFLVSKGRALMPSLFDEEITAGYAGLRAATDHDDYLIDVDVQQRYLLVGGIRSTGLTSGMAI AEYVSGLLGEAGVDVTERDDLPPPPRMPNLGEAGVRPYQDTGRVAADPEYGRIVCFCERVSAGEIRDAFD SPIPPADLDGLRRRTRVMNGRCQGFYCGAHTQALLRAGADR
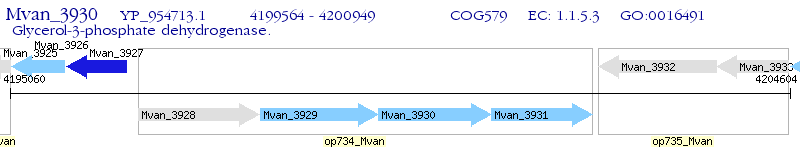
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_3930 | - | - | 100% (461) | FAD dependent oxidoreductase |
| M. vanbaalenii PYR-1 | Mvan_0204 | - | 8e-23 | 26.79% (392) | hypothetical protein Mvan_0204 |
| M. vanbaalenii PYR-1 | Mvan_2290 | - | 3e-10 | 23.31% (369) | FAD dependent oxidoreductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2273c | glpD1 | 8e-14 | 27.85% (395) | glycerol-3-phosphate dehydrogenase |
| M. gilvum PYR-GCK | Mflv_2649 | - | 0.0 | 87.31% (457) | FAD dependent oxidoreductase |
| M. tuberculosis H37Rv | Rv2249c | glpD1 | 8e-14 | 27.85% (395) | glycerol-3-phosphate dehydrogenase |
| M. leprae Br4923 | MLBr_00713 | glpD | 1e-06 | 23.88% (402) | putative glycerol-3-phosphate dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3882c | - | 3e-15 | 28.03% (346) | glycerol-3-phosphate dehydrogenase 1 GlpD1 |
| M. marinum M | MMAR_3342 | glpD1 | 1e-12 | 27.62% (391) | glycerol-3-phosphate dehydrogenase GlpD1 |
| M. avium 104 | MAV_2188 | - | 8e-13 | 28.06% (392) | glycerol-3-phosphate dehydrogenase 1 |
| M. smegmatis MC2 155 | MSMEG_1379 | - | 8e-18 | 26.49% (385) | hypothetical protein MSMEG_1379 |
| M. thermoresistible (build 8) | TH_4606 | - | 2e-13 | 28.17% (394) | PUTATIVE FAD dependent oxidoreductase |
| M. ulcerans Agy99 | MUL_1300 | glpD1 | 4e-13 | 27.62% (391) | glycerol-3-phosphate dehydrogenase GlpD1 |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_3930|M.vanbaalenii_PYR-1 -----------------------------MSRQDISDVVVIGAGIVGCAI
Mflv_2649|M.gilvum_PYR-GCK -----------------------------MRPEDVSDVVVIGAGIVGCAI
Mb2273c|M.bovis_AF2122/97 ---------MLMPHSAALNAARRSADLTALADGGALDVIVIGGGITGVGI
Rv2249c|M.tuberculosis_H37Rv ---------MLMPHSAALNAARRSADLTALADGGALDVIVIGGGITGVGI
MMAR_3342|M.marinum_M -----------MPDSTALNAARRSTELTALADGEALDVLVIGGGITGVGI
MUL_1300|M.ulcerans_Agy99 -----------MPDSTALNAARRSTELTALADGEALDVLVIGGGITGVGI
MAV_2188|M.avium_104 ----------MIPDPSALNAARRTADLAALAEGEPLDVLVIGGGITGAGI
TH_4606|M.thermoresistible__bu -------VSAKKGDATALNASRRAAELTALAEQR-LDILVIGGGITGAGI
MAB_3882c|M.abscessus_ATCC_199 -------------MTTQLNSTRRSADLDALSGGAPIDVLVIGGGITGVGV
MLBr_00713|M.leprae_Br4923 MTDLIQAPESEQTWPASALGPQQRAAAWERFGTEQFDVVVIGGGVVGSGC
MSMEG_1379|M.smegmatis_MC2_155 --------------------------------MAAPRIGVIGAGIVGLAV
: ***.*:.* .
Mvan_3930|M.vanbaalenii_PYR-1 ARALSGT-GLSVTLLEARGDVGDGTSKANTALLHTGFDATPG---TLESR
Mflv_2649|M.gilvum_PYR-GCK ARALAGT-HLSVTLVEARDDVGDGTSKANTALLHTGFDATPG---TLESR
Mb2273c|M.bovis_AF2122/97 ALDAATR-GLTVALVEKH-DLAFGTSRWSSKLVHGGLRYLASGNVGIARR
Rv2249c|M.tuberculosis_H37Rv ALDAATR-GLTVALVEKH-DLAFGTSRWSSKLVHGGLRYLASGNVGIARR
MMAR_3342|M.marinum_M ALDAASR-GLQVALVEKH-DLAFGTSRWSSKLVHGGLRYLASGNVGIARR
MUL_1300|M.ulcerans_Agy99 ALDAASR-GLQVALVEKH-DLAFGTSRWSSKLVHGGLRYLASGNVGIARR
MAV_2188|M.avium_104 ALDAAAR-GLRVALVEKR-DLAFGTSRWSSKLVHGGLRYLASGNVGIARR
TH_4606|M.thermoresistible__bu ALDAASR-GLTVALVEKH-DLAFGTSRWSSKLVHGGLRYLASGNIGIARR
MAB_3882c|M.abscessus_ATCC_199 ALDAASR-GLRVTLVEKH-DLAFGTSRFSSKLVHGGLRYLASGQVGVARE
MLBr_00713|M.leprae_Br4923 ALDAATR-GLKVALVEAR-DLASGTSSRSSKMFHGGLRYLEQLEFGLVRE
MSMEG_1379|M.smegmatis_MC2_155 ARRLQQKLQADVTVLDKENVVAAHQTGHNSNVVHSGVYYPPG---SLKAT
* *:::: . :. : .: :.* *. :
Mvan_3930|M.vanbaalenii_PYR-1 LVARGYQLLGEYAGQTGIPVERTGAMLVAWTEEEAGALPGLMVKAERNG-
Mflv_2649|M.gilvum_PYR-GCK LVARGYHLLGEYAEQTGIPVERTGAMLVAWTDEEADALPGLMVKAERNG-
Mb2273c|M.bovis_AF2122/97 SAVERGILMTRNAPHLVHAMPQLVPLLPSMGHTKRALVRAGFLAGDALRV
Rv2249c|M.tuberculosis_H37Rv SAVERGILMTRNAPHLVHAMPQLVPLLPSMGHTKRALVRAGFLAGDALRV
MMAR_3342|M.marinum_M SAIERGILMTRNAPHLVHAMPQLVPLLPSMDHLKRALVRTGFLAGDALRM
MUL_1300|M.ulcerans_Agy99 SAIERGILMTRNAPHLVHAMPQLVPLLPSMDHLKRALVRTGFLAGDALRM
MAV_2188|M.avium_104 SALERGILMTRNAPHLVHAMPQLVPLLPSMSRATRALVRGGFLAGDALRF
TH_4606|M.thermoresistible__bu SAVERGILMTRNAPHLVKAMPQLVPLIPTIGTLERALVRTGFLAGDVLRR
MAB_3882c|M.abscessus_ATCC_199 SAVERHILMTRTAPHLIHPLAQVVPLHPAVSMFSRTLVRVGFIAGDGLRK
MLBr_00713|M.leprae_Br4923 ALYERELSLTTLAPHLVKPLPFLFPLT--KRWWERPYIAAGIFLYDRLG-
MSMEG_1379|M.smegmatis_MC2_155 LCRRGVGLLRAYCMERGLPYDELGKVIVAVQTDELPRLHDLAKRAVDNG-
. : . . . : :
Mvan_3930|M.vanbaalenii_PYR-1 --------YRACGIVGADEVYRRVPDLGPGALAG-LDVPGEAIICTWTTN
Mflv_2649|M.gilvum_PYR-GCK --------YRDCDLVRSEEVYRRVPDLGLGALGG-LGVPGESIICTWTTN
Mb2273c|M.bovis_AF2122/97 LAGTPAATLPRSRRIPASRVVEIAPTVRRDGLDGGLLAYDGQLIDDARLV
Rv2249c|M.tuberculosis_H37Rv LAGTPAATLPRSRRIPASRVVEIAPTVRRDGLDGGLLAYDGQLIDDARLV
MMAR_3342|M.marinum_M LAGTPSSTLPRSRRVSAQRVVELAPTVRRDGLAGGLLAYDGQLIDDARLV
MUL_1300|M.ulcerans_Agy99 LAGTPSSTLPRSRRVSAQRVVELAPTVRRDGLAGGLLAYDGQLIDDARLV
MAV_2188|M.avium_104 LAGTPASTLPRSQRVSAQRVVQLAPTVCRDGLDGGLLAYDGQLIDDARLV
TH_4606|M.thermoresistible__bu LAGTPASTLPRSGRVDAGRAAELVPAVRRDRLDGALLAYDGQLIDDARLV
MAB_3882c|M.abscessus_ATCC_199 LAHTSAEVLPRSRAINAGEVIRRAPTVRRNGLRGGLMAFDGQLVDDARLV
MLBr_00713|M.leprae_Br4923 ----GAKSVPAQKHLTRAGALRLSPGLKRSSLIGGIRYYD-TVVDDARHT
MSMEG_1379|M.smegmatis_MC2_155 --------IPDTRVIDRAELREREPHVDG---LAALLIPSTAVVSFPAIA
: . * : . : . ::
Mvan_3930|M.vanbaalenii_PYR-1 LALATDAVARGARLLRGARVTAVETGP-------EHTTLRTTSGEVRARW
Mflv_2649|M.gilvum_PYR-GCK LALATDAVARGARLLRGARVTGVQPGP-------EHTTLHTTAGDVRGRW
Mb2273c|M.bovis_AF2122/97 MAVARTAAQHGARILTYVGASNVTGTS-----VELTDRRTRQSFALSARA
Rv2249c|M.tuberculosis_H37Rv MAVARTAAQHGARILTYVGASNVTGTS-----VELTDRRTRQSFALSARA
MMAR_3342|M.marinum_M AAVARTAAQYGARILTYVGASEATGTS-----ARLTDQRSGESFDVSARA
MUL_1300|M.ulcerans_Agy99 TAVARTAAQHGARILTYVGASEATGTS-----ARLTDQRSGESFDVSARA
MAV_2188|M.avium_104 TAVARTAAQHGARILTYVAASRATGTS-----VRLTDVRGGGSFDVSAGA
TH_4606|M.thermoresistible__bu TAVARTAAQHGARILTRVAADRARGDG-----AALTDQLTGESFDVAARA
MAB_3882c|M.abscessus_ATCC_199 VSIARTAAGLGARILTRVAAEQVTGDG-----AVLRDRLTGNAIRVRAGL
MLBr_00713|M.leprae_Br4923 LTVARTAAHYGAVVRCSTQVVALLREGDRVIGVRVRDSEDGAVTEIRGHV
MSMEG_1379|M.smegmatis_MC2_155 EEFRADIEAAGGQFRLGSPAVGIASRD-------GGVVITTDTDEFVFDH
. *. . . .
Mvan_3930|M.vanbaalenii_PYR-1 VINAAGLGADHLDAEFGYRRFTVTPRRGELLVFDKLTR-PMVPLIVLAVP
Mflv_2649|M.gilvum_PYR-GCK IINAAGLGADHLDTEFGHRRFTVTPRRGELLVFDKLTR-PMVPLIVLAVP
Mb2273c|M.bovis_AF2122/97 VINAAGVWAGEIDPSLRLR-----PSRGTHLVFDAKSFANPTAALTIPIP
Rv2249c|M.tuberculosis_H37Rv VINAAGVWAGEIDPSLRLR-----PSRGTHLVFDAKSFANPTAALTIPIP
MMAR_3342|M.marinum_M VINAAGVWGGQIDPALKLR-----PSRGTHLVFDANAFGNPTAALTIPIP
MUL_1300|M.ulcerans_Agy99 VINAAGVWGGQIDPALKLR-----PSRGTHLVFDANAFGNPTAALTIPIP
MAV_2188|M.avium_104 VINAAGVWAGEIDGSLRLR-----PSRGTHLVFDAAAFGNPTAALTIPIP
TH_4606|M.thermoresistible__bu VINATGVWAGQVDPALRLR-----PSRGTHLVFDAAAFGNPVAALTIPIP
MAB_3882c|M.abscessus_ATCC_199 VINAAGVWAGQVDREIKLR-----PSRGTHLVFRAASFGGLSAALTVPVP
MLBr_00713|M.leprae_Br4923 VVNATGVWTDEIQALSKQR-----GRFQVRVSKGVHVVVPRDRIVSDVAM
MSMEG_1379|M.smegmatis_MC2_155 VVVCAGLQSSMIAEMAGAP-------RDPEIIPFRGEYYSLVASRSDLVK
:: .:*: . : :
Mvan_3930|M.vanbaalenii_PYR-1 SSRGKGVLVSPTIYGNVMVGPTSENLEDRTATG--TSEKGFEFLVSKG-R
Mflv_2649|M.gilvum_PYR-GCK SSRGKGVLVSPTIYGNVMVGPTSENLDDRTATG--TSEHGFEFLVSKG-R
Mb2273c|M.bovis_AF2122/97 GELNRFVFAMPEQLGRIYLGLTDEDAPGPIPDVPQPSSEEITFLLDTVNT
Rv2249c|M.tuberculosis_H37Rv GELNRFVFAMPEQLGRIYLGLTDEDAPGPIPDVPQPSSEEITFLLDTVNT
MMAR_3342|M.marinum_M GELNRFVFAMPEQLGRVYLGLTDEDAPGPIPDVPEPSSEEIMFLLDTVNT
MUL_1300|M.ulcerans_Agy99 GELNRFVFAMPEQLGRVYLGLTDEDAPGPFLDVPEPSSEEIMFLLDTVNT
MAV_2188|M.avium_104 GELNRFVFAMPEQLGRVYLGLTDEAAPGPIPDVPEPSVEEITFLLDTVNT
TH_4606|M.thermoresistible__bu GELNRFVFAMPEQLGRVYLGLTDEDAPGPIPDVPQPTPQEVSFLLDTVNT
MAB_3882c|M.abscessus_ATCC_199 GTINRFVFALPAIHDRVYLGLTDEPAPGPVPDVAEPSAAEERFLLDTANT
MLBr_00713|M.leprae_Br4923 ILRTKKSVMFIIPWGNHWIIGTTDTDWNLDLAHPAATKADIDYILQTVNT
MSMEG_1379|M.smegmatis_MC2_155 GLVYPVPDPRYPFLGVHFTRGIDGHVHVGPNAVLALAQEGYRWRDVNVRQ
. : : .
Mvan_3930|M.vanbaalenii_PYR-1 ALMPSLFDEEITAGYAGLRAATDHDD---YLIDVDVQQRYLLVGGIRSTG
Mflv_2649|M.gilvum_PYR-GCK ALMPALFEEEVTATYAGLRAAADHDD---YLIDLDERLRYVLVGGIRSTG
Mb2273c|M.bovis_AF2122/97 ALGTAVGTKDVIGAYAGLRPLIDTGGAGVQGRTADVSRDHAVFESPSGVI
Rv2249c|M.tuberculosis_H37Rv ALGTAVGTKDVIGAYAGLRPLIDTGGAGVQGRTADVSRDHAVFESPSGVI
MMAR_3342|M.marinum_M ALGTALQTSDVIGAYAGLRPLIDTG----EGRTADVSRDHAVIESPSGVI
MUL_1300|M.ulcerans_Agy99 ALGTALQTSDVIGAYAGLRPLIDTG----EGRTADVSRDHAVIESPSGVI
MAV_2188|M.avium_104 ALGTALAATDVIGAYAGLRPLIDTG----EGRTADVSREHAVVESASGVI
TH_4606|M.thermoresistible__bu ALATALGPDDVIGAYAGLRPLIDTP----GGRTADISRDHAVVESDTGLF
MAB_3882c|M.abscessus_ATCC_199 VLQTSLSETDVVGRFAGLRPLLDGD----DSATADLSRKHAVRVSESGVV
MLBr_00713|M.leprae_Br4923 VLATPLTHADIDGVYAGLRPLLAGE----SDDTSKLTREHAVAVPVAGLV
MSMEG_1379|M.smegmatis_MC2_155 LWRSVS--------YPGMLRLARHH-------------------------
. :.*:
Mvan_3930|M.vanbaalenii_PYR-1 LTSGMAIAEYVSGLLGEAGVDVTERDDLPPPPRMPNLGEAGVRPY-----
Mflv_2649|M.gilvum_PYR-GCK LTSGMAVAEYVSGLLARAGVAVDERDDLPPPPRMPNIGEAGLRPY-----
Mb2273c|M.bovis_AF2122/97 SVVGGKLTEYRYMAEDVLNRAITLRHLRAAKCRTRNLPLIGAPANPGPAP
Rv2249c|M.tuberculosis_H37Rv SVVGGKLTEYRYMAEDVLNRAITLRHLRAAKCRTRNLPLIGAPANPGPAP
MMAR_3342|M.marinum_M SVVGGKLTEYRYMAEDVLNRAISLRQLQANQCRTRDLPLIGAPANPGPAA
MUL_1300|M.ulcerans_Agy99 SVVGGKLTEYRYMAEDVLNRAISLRQLQATQCRTRDLPLIGAPATPGPAA
MAV_2188|M.avium_104 SVIGGKLTEYRYMAQDVLDRAVRVRRLRAAKCRTRNLPLIGAPANPGITG
TH_4606|M.thermoresistible__bu SVLGGKLTEYRHMAEDAVDRAVTSRGLRAGECRTRNLPLVGAPANPVATF
MAB_3882c|M.abscessus_ATCC_199 SVVGGKLTTYRKMAQDALDAGLQRRPLAASECVTQNLAVVDDWP------
MLBr_00713|M.leprae_Br4923 AIAGGKYTTYRVMAADAIDAAVAFVPARVAPSITEKVGLLGADGYFALIN
MSMEG_1379|M.smegmatis_MC2_155 ---------WRMGASEIVGSMSKSVFLRRARAYIPELTAS----------
: . .:
Mvan_3930|M.vanbaalenii_PYR-1 ------------------------QDTGRVAADPEYG----RIVCFCERV
Mflv_2649|M.gilvum_PYR-GCK ------------------------QDADRVAADPEYG----RIVCFCERV
Mb2273c|M.bovis_AF2122/97 GSGA---------GLPESLVARYGAEAANVAAAATCE-RPTEPVADGIDV
Rv2249c|M.tuberculosis_H37Rv GSGA---------GLPESLVARYGAEAANVAAAATCE-RPTEPVADGIDV
MMAR_3342|M.marinum_M GPG----------GLPESLVARFGAEAANVVATATCE-RPTDPVAEGIDV
MUL_1300|M.ulcerans_Agy99 GPG----------GLPEFLVARFGAEAANVVVTATCE-RPTDPVAEGIDV
MAV_2188|M.avium_104 APQV---------ALPESLVSRYGAEAAKVVATAGCE-RPSEPVVDGIDV
TH_4606|M.thermoresistible__bu RSAVDSGAE-LPARLPSSLVARYGAEAPNVIARAACD-RPTDPVAEGIDV
MAB_3882c|M.abscessus_ATCC_199 -----------------------GAAGAG--------------LVEGVDI
MLBr_00713|M.leprae_Br4923 QVEHVAALQGLHPYRVRHLLDRYGALIGDVLALAAEAPDLLSPIQEAPGY
MSMEG_1379|M.smegmatis_MC2_155 ---------------------------------------------DIVRA
Mvan_3930|M.vanbaalenii_PYR-1 SAGEIRDAFDSPIPPADLDGLRRRTRVMNGRCQG-----FYCGAHTQALL
Mflv_2649|M.gilvum_PYR-GCK TAGEIRDAFGSLIPPADLDGLRRRTRVMNGRCQG-----FYCGAHTEALL
Mb2273c|M.bovis_AF2122/97 TRAEFEYAVTHEGALDVDDILDRRTRIGLVPRDR-----ERVVAVAKEFL
Rv2249c|M.tuberculosis_H37Rv TRAEFEYAVTHEGALDVDDILDRRTRIGLVPRDR-----ERVVAVAKEFL
MMAR_3342|M.marinum_M TRAEFEYAVTHEGALDAEDILDRRTRIGLVPHDR-----QRVVGIAEEFL
MUL_1300|M.ulcerans_Agy99 TRAEFEYAVTHEGALDAEDILDRRTRIGLVPHDR-----ERVVGIAEEFL
MAV_2188|M.avium_104 TRAEFEYAITHEGALEVSDIVDRRTRIGLVPRDR-----ERVVAVAEEFL
TH_4606|M.thermoresistible__bu IRAEFEYAVTHEGAMTVEDILDRRTRIGLVAADR-----QRAVAAAEEIL
MAB_3882c|M.abscessus_ATCC_199 TREQVEFALTHEGALDADDILDRRTRIGLVPVDK-----NQAIGAVQRLV
MLBr_00713|M.leprae_Br4923 LKVEARYAVTAEGALHLEDILARRMRVSIEYPHRGVACAREVADVVAPVL
MSMEG_1379|M.smegmatis_MC2_155 ESGVRAQALKPDGNLVDDFVIHRLAGMTFVRNAP-----SPAATASLAIA
*. : * : .
Mvan_3930|M.vanbaalenii_PYR-1 RAGADR--------------------------------------------
Mflv_2649|M.gilvum_PYR-GCK EALLETGALR----------------------------------------
Mb2273c|M.bovis_AF2122/97 SR------------------------------------------------
Rv2249c|M.tuberculosis_H37Rv SR------------------------------------------------
MMAR_3342|M.marinum_M AGVG----------------------------------------------
MUL_1300|M.ulcerans_Agy99 AGVG----------------------------------------------
MAV_2188|M.avium_104 ARFG----------------------------------------------
TH_4606|M.thermoresistible__bu AAAE----------------------------------------------
MAB_3882c|M.abscessus_ATCC_199 DSYFSQVN------------------------------------------
MLBr_00713|M.leprae_Br4923 GWTAEDIDREVATYNARVEAEVLSQAQPDDVSADMLRASAPEARTKIIEP
MSMEG_1379|M.smegmatis_MC2_155 EHIVDALA------------------------------------------
Mvan_3930|M.vanbaalenii_PYR-1 ----
Mflv_2649|M.gilvum_PYR-GCK ----
Mb2273c|M.bovis_AF2122/97 ----
Rv2249c|M.tuberculosis_H37Rv ----
MMAR_3342|M.marinum_M ----
MUL_1300|M.ulcerans_Agy99 ----
MAV_2188|M.avium_104 ----
TH_4606|M.thermoresistible__bu ----
MAB_3882c|M.abscessus_ATCC_199 ----
MLBr_00713|M.leprae_Br4923 VSLT
MSMEG_1379|M.smegmatis_MC2_155 ----