For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
KGRVGPRIAVIAAILTMVVGAAGFFVTLMLNAFVFDEFDAYGEVPVPGSGRLHLPAGEVTISFHTAVTGG TGGGFPVPALQLAISPSAGGPDPVVTETMGSTTSVNNDVRVRIWVAQIPADGTYDIETGGNVRGYIDPRL AFGRDTSSGWLPWALGGLFVAGLAGLAPALVWSSRAGRRPQPLRGPVSLDEPSWPGPLPPPVASYQPTDQ GVRLEQLKTLAALRDSGALTAAEFEAEKRRILDG*
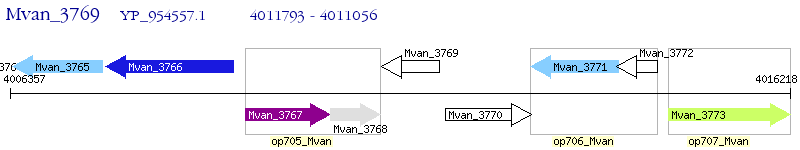
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_3769 | - | - | 100% (245) | hypothetical protein Mvan_3769 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_3353 | - | 2e-72 | 54.40% (250) | hypothetical protein MMAR_3353 |
| M. avium 104 | MAV_2746 | - | 2e-05 | 27.44% (215) | PPE family protein |
| M. smegmatis MC2 155 | MSMEG_4343 | - | 3e-66 | 50.78% (256) | hypothetical protein MSMEG_4343 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_1290 | - | 8e-72 | 54.00% (250) | hypothetical protein MUL_1290 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_3353|M.marinum_M --------------------------------------------------
MUL_1290|M.ulcerans_Agy99 --------------------------------------------------
Mvan_3769|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4343|M.smegmatis_MC2_155 --------------------------------------------------
MAV_2746|M.avium_104 MDFAVLPPEVNSARMYAGPGSGPMLAAAMAWDELAATLHSTADSYQAEIT
MMAR_3353|M.marinum_M --------------------------------------------------
MUL_1290|M.ulcerans_Agy99 --------------------------------------------------
Mvan_3769|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4343|M.smegmatis_MC2_155 --------------------------------------------------
MAV_2746|M.avium_104 ALTSGPWVGPSAAAMIAAITPYLTWMRTTGAQAEETANNARAAAFAYETA
MMAR_3353|M.marinum_M ---------MTS-RRVAKFSLTVAILTLVVSVAGFIVTLVLNAFFLDKYD
MUL_1290|M.ulcerans_Agy99 ---------MTN-RRVAKFSLTVAILTLVVSVAGFIVTLVLNAFFLDKYD
Mvan_3769|M.vanbaalenii_PYR-1 ---------MKG-RVGPRIAVIAAILTMVVGAAGFFVTLMLNAFVFDEFD
MSMEG_4343|M.smegmatis_MC2_155 ---------MNSSRAIPRVAVVLSGCAVVIGIVGFVVALILNALFFDKFD
MAV_2746|M.avium_104 FAETVPPPVVTANRTLLAQLVATNILGQNTPAIATTEAEYAEMWARDAAA
:. * : . : : *
MMAR_3353|M.marinum_M AYGEVAIPGSGSVHLPAG----------DVTVSLHTLVIGGTNGVGLPVP
MUL_1290|M.ulcerans_Agy99 AYGEVAIPGSGGVHLPAG----------DVTVSLHTLVIGGTNGVGLPVP
Mvan_3769|M.vanbaalenii_PYR-1 AYGEVPVPGSGRLHLPAG----------EVTISFHTAVTGGTGG-GFPVP
MSMEG_4343|M.smegmatis_MC2_155 AYGEVPIPGSGSVELPAG----------EVSVSFHTWVTDSEGG--LPVP
MAV_2746|M.avium_104 MYGYAGAAAAATRLTPFTQPDPTTNSGADQSQAVSKAAATAAGSTRNTVT
** . ..:. * : : :. . . . .. .*.
MMAR_3353|M.marinum_M PLGISISP---------PDGVPQPEVTESI--------------------
MUL_1290|M.ulcerans_Agy99 PLGISISP---------PDGVPQPEVTESI--------------------
Mvan_3769|M.vanbaalenii_PYR-1 ALQLAISP---------SAGGPDPVVTETM--------------------
MSMEG_4343|M.smegmatis_MC2_155 ALELHVQA---------PPGAAMPTVTESP--------------------
MAV_2746|M.avium_104 QQPFSAVPNMLNNLAVSPNALGDPSVTDLLGLESDLIAIFLDAPASVVGM
: . . . * **:
MMAR_3353|M.marinum_M ----------------GSTTTVNNDAHVRVWMVRVPVEATYRVTTDGEVG
MUL_1290|M.ulcerans_Agy99 ----------------GSTTTVNNDAHVRVWMVRVPVEATYRVTTDGEVG
Mvan_3769|M.vanbaalenii_PYR-1 ----------------GSTTSVNNDVRVRIWVAQIPADGTYDIETGGNVR
MSMEG_4343|M.smegmatis_MC2_155 ----------------GATTTVNNDAHVRVWMMQVAEPAVYQVRVDGDVR
MAV_2746|M.avium_104 GVDAPSATIALPFDIAGALTGFHTDDIVSGWAGVQPWPGTGAVPPS-PFP
*: * .:.* * * . .. : . .
MMAR_3353|M.marinum_M GYINPRLAFGHQSSYGWLVWVFVAMFGVG--------------LIDLVLS
MUL_1290|M.ulcerans_Agy99 GYINPRLAYGHQSSYGWLVWVFVAMFGVG--------------LIDLVLS
Mvan_3769|M.vanbaalenii_PYR-1 GYIDPRLAFGRDTSSGWLPWALGGLFVAG--------------LAGLAPA
MSMEG_4343|M.smegmatis_MC2_155 GYINPRLAFGYPSRHWWLVWMFAGVFAAS--------------LAALAAS
MAV_2746|M.avium_104 VITNPAGGFGTLASAGLGQAKTVGTLSVPPTWTAAAPAIRPAALALPATT
:* .:* : . : . * . :
MMAR_3353|M.marinum_M VVWLSRS--RPAASAS-----PASPHFAEAPVLASHPSDPPRTAYEPSDD
MUL_1290|M.ulcerans_Agy99 VVWLSRS--RPAASAS-----PASPYFAEAPVLASHPSDPPRTAYEPSDD
Mvan_3769|M.vanbaalenii_PYR-1 LVWSSRAGRRPQPLRG-----PVS---LDEPSWPG-PLPPPVASYQPTDQ
MSMEG_4343|M.smegmatis_MC2_155 AMWLRRAERRPRFLGSGERFEPVEAVELGEHTINIGGPSERGYSYQPTDH
MAV_2746|M.avium_104 VTAAAETAATAAPAGTGSLFGEMAAAGLAGRAMAGTTGLGRERARAEAAK
.: . : : .
MMAR_3353|M.marinum_M GIR----------------------------LERLKTLAALRDSGALTEE
MUL_1290|M.ulcerans_Agy99 GIR----------------------------LERLKTLAALRDSGALTEE
Mvan_3769|M.vanbaalenii_PYR-1 GVR----------------------------LEQLKTLAALRDSGALTAA
MSMEG_4343|M.smegmatis_MC2_155 AIR----------------------------IEQLKTIAALRDSGALTQE
MAV_2746|M.avium_104 APREPGQEPGKAPKQEPAAPAVSAGGPITSIAAELRELASLRDAGILTQQ
. * .*: :*:***:* **
MMAR_3353|M.marinum_M EFQSEKRRILDGR
MUL_1290|M.ulcerans_Agy99 EFQSEKRRILDGR
Mvan_3769|M.vanbaalenii_PYR-1 EFEAEKRRILDG-
MSMEG_4343|M.smegmatis_MC2_155 EFEAEKRRILDS-
MAV_2746|M.avium_104 EFEEQKKRLLPS-
**: :*:*:* .