For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TSRRVAKFSLTVAILTLVVSVAGFIVTLVLNAFFLDKYDAYGEVAIPGSGSVHLPAGDVTVSLHTLVIGG TNGVGLPVPPLGISISPPDGVPQPEVTESIGSTTTVNNDAHVRVWMVRVPVEATYRVTTDGEVGGYINPR LAFGHQSSYGWLVWVFVAMFGVGLIDLVLSVVWLSRSRPAASASPASPHFAEAPVLASHPSDPPRTAYEP SDDGIRLERLKTLAALRDSGALTEEEFQSEKRRILDGR*
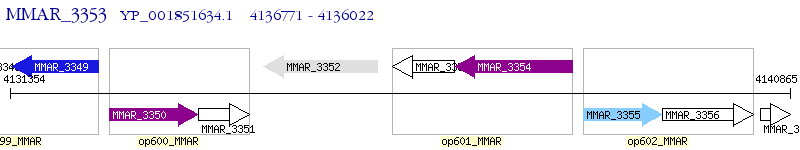
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_3353 | - | - | 100% (249) | hypothetical protein MMAR_3353 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_4343 | - | 2e-67 | 51.97% (254) | hypothetical protein MSMEG_4343 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_1290 | - | 1e-139 | 98.39% (249) | hypothetical protein MUL_1290 |
| M. vanbaalenii PYR-1 | Mvan_3769 | - | 2e-72 | 54.40% (250) | hypothetical protein Mvan_3769 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_3353|M.marinum_M MTS-RRVAKFSLTVAILTLVVSVAGFIVTLVLNAFFLDKYDAYGEVAIPG
MUL_1290|M.ulcerans_Agy99 MTN-RRVAKFSLTVAILTLVVSVAGFIVTLVLNAFFLDKYDAYGEVAIPG
Mvan_3769|M.vanbaalenii_PYR-1 MKG-RVGPRIAVIAAILTMVVGAAGFFVTLMLNAFVFDEFDAYGEVPVPG
MSMEG_4343|M.smegmatis_MC2_155 MNSSRAIPRVAVVLSGCAVVIGIVGFVVALILNALFFDKFDAYGEVPIPG
*.. * .:.:: : ::*:. .**.*:*:***:.:*::******.:**
MMAR_3353|M.marinum_M SGSVHLPAGDVTVSLHTLVIGGTNGVGLPVPPLGISISPPDGVPQPEVTE
MUL_1290|M.ulcerans_Agy99 SGGVHLPAGDVTVSLHTLVIGGTNGVGLPVPPLGISISPPDGVPQPEVTE
Mvan_3769|M.vanbaalenii_PYR-1 SGRLHLPAGEVTISFHTAVTGGTGG-GFPVPALQLAISPSAGGPDPVVTE
MSMEG_4343|M.smegmatis_MC2_155 SGSVELPAGEVSVSFHTWVTDSEGG--LPVPALELHVQAPPGAAMPTVTE
** :.****:*::*:** * .. .* :***.* : :... * . * ***
MMAR_3353|M.marinum_M SIGSTTTVNNDAHVRVWMVRVPVEATYRVTTDGEVGGYINPRLAFGHQSS
MUL_1290|M.ulcerans_Agy99 SIGSTTTVNNDAHVRVWMVRVPVEATYRVTTDGEVGGYINPRLAYGHQSS
Mvan_3769|M.vanbaalenii_PYR-1 TMGSTTSVNNDVRVRIWVAQIPADGTYDIETGGNVRGYIDPRLAFGRDTS
MSMEG_4343|M.smegmatis_MC2_155 SPGATTTVNNDAHVRVWMMQVAEPAVYQVRVDGDVRGYINPRLAFGYPSR
: *:**:****.:**:*: ::. ..* : ..*:* ***:****:* :
MMAR_3353|M.marinum_M YGWLVWVFVAMFGVGLIDLVLSVVWLSRS--RPAASAS-----PASPHFA
MUL_1290|M.ulcerans_Agy99 YGWLVWVFVAMFGVGLIDLVLSVVWLSRS--RPAASAS-----PASPYFA
Mvan_3769|M.vanbaalenii_PYR-1 SGWLPWALGGLFVAGLAGLAPALVWSSRAGRRPQPLRG-----PVS---L
MSMEG_4343|M.smegmatis_MC2_155 HWWLVWMFAGVFAASLAALAASAMWLRRAERRPRFLGSGERFEPVEAVEL
** * : .:* ..* *. : :* *: ** . *..
MMAR_3353|M.marinum_M EAPVLASHPSDPPRTAYEPSDDGIRLERLKTLAALRDSGALTEEEFQSEK
MUL_1290|M.ulcerans_Agy99 EAPVLASHPSDPPRTAYEPSDDGIRLERLKTLAALRDSGALTEEEFQSEK
Mvan_3769|M.vanbaalenii_PYR-1 DEPSWPG-PLPPPVASYQPTDQGVRLEQLKTLAALRDSGALTAAEFEAEK
MSMEG_4343|M.smegmatis_MC2_155 GEHTINIGGPSERGYSYQPTDHAIRIEQLKTIAALRDSGALTQEEFEAEK
:*:*:*..:*:*:***:********** **::**
MMAR_3353|M.marinum_M RRILDGR
MUL_1290|M.ulcerans_Agy99 RRILDGR
Mvan_3769|M.vanbaalenii_PYR-1 RRILDG-
MSMEG_4343|M.smegmatis_MC2_155 RRILDS-
*****.