For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
KAPVAQQQLLADLAELDAEVSRNEHRTKNLPEQKAVEEAQAAHREASDRLAALQLALADIDAQVAKLESE IDGVRQREDRDRALLDGGTVDPKQLTDLQHELDTLQRRQASLEDSQLEVMERREELAAEQTREQAAIEEL QTALTGAQRACDDARAELAQAREKSAARRAELVSAIDGELVALYERQRARGGVGAGVLQGRRCGACRIEI DQGESARIAAAAEDEVLRCPECSAILLRVKGSRR*
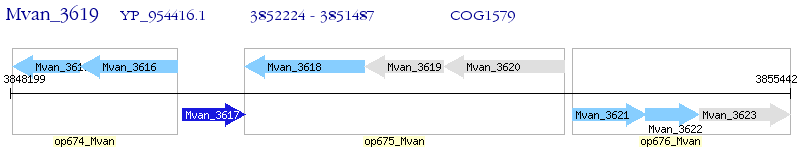
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_3619 | - | - | 100% (245) | hypothetical protein Mvan_3619 |
| M. vanbaalenii PYR-1 | Mvan_2175 | - | 2e-07 | 28.48% (158) | chromosome segregation protein SMC |
| M. vanbaalenii PYR-1 | Mvan_4435 | - | 2e-06 | 28.08% (203) | hypothetical protein Mvan_4435 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2254c | - | 3e-78 | 59.92% (242) | hypothetical protein Mb2254c |
| M. gilvum PYR-GCK | Mflv_2891 | - | 1e-105 | 79.10% (244) | hypothetical protein Mflv_2891 |
| M. tuberculosis H37Rv | Rv2229c | - | 1e-78 | 60.33% (242) | hypothetical protein Rv2229c |
| M. leprae Br4923 | MLBr_01638 | - | 3e-54 | 50.22% (229) | hypothetical protein MLBr_01638 |
| M. abscessus ATCC 19977 | MAB_1904 | - | 1e-67 | 55.97% (243) | hypothetical protein MAB_1904 |
| M. marinum M | MMAR_3305 | - | 5e-81 | 63.22% (242) | hypothetical protein MMAR_3305 |
| M. avium 104 | MAV_2210 | - | 2e-80 | 61.98% (242) | hypothetical protein MAV_2210 |
| M. smegmatis MC2 155 | MSMEG_4306 | - | 6e-85 | 67.22% (241) | hypothetical protein MSMEG_4306 |
| M. thermoresistible (build 8) | TH_1466 | - | 2e-83 | 63.90% (241) | conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_1323 | - | 1e-80 | 62.81% (242) | hypothetical protein MUL_1323 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2254c|M.bovis_AF2122/97 MKAGVAQQRSLLELAKLDAELTRIAHRATHLPQRAAYQQVQAEHNAANDR
Rv2229c|M.tuberculosis_H37Rv MKAGVAQQRSLLELAKLDAELTRIAHRATHLPQRAAYQQVQAEHNAANDR
MMAR_3305|M.marinum_M MKAEAAQQRSLLELAKLDAELSRIAHRSTHLPEGSAFEQVRVQHEAVSDR
MUL_1323|M.ulcerans_Agy99 MKAEAAQQRSLLELAKLDAELSRIAHRSTHLPEGSAFEQVRVQHEAVSDR
Mvan_3619|M.vanbaalenii_PYR-1 MKAPVAQQQLLADLAELDAEVSRNEHRTKNLPEQKAVEEAQAAHREASDR
Mflv_2891|M.gilvum_PYR-GCK MKAAVAQQQLLLELAEVDAEISRVEHRTKNLPEQKAVEEAQAALREVGDR
MSMEG_4306|M.smegmatis_MC2_155 MKAEVSQQRSLLTLSEVDAELARIAHRGKNLAEQKRLDELTAQRGEVNDR
TH_1466|M.thermoresistible__bu ----VDQQRLLLELAEIDAELARIDHRTKNLPARAELEAQQRRHQEATDR
MAB_1904|M.abscessus_ATCC_1997 MKADVAQQRLLVDLASVDAELTRVAHRRANPPERQEHGELQAQQRTILDE
MAV_2210|M.avium_104 MKADVAQQRSLLELANVDAELSRLAHRAEHLPEQQACERMQQEYDAAGDR
MLBr_01638|M.leprae_Br4923 -------------MSKLDDELSRIAHRANYLPQREAYERMRVERTGANDR
::.:* *::* ** . *.
Mb2254c|M.bovis_AF2122/97 MAALRIAAEDLDGQVSRFESEIDAVRKRGDRDRSLLTSGATDAKQLADLQ
Rv2229c|M.tuberculosis_H37Rv MAALRIAAEDLDGQVSRFESEIDAVRKRGDRDRSLLTSGATDAKQLADLQ
MMAR_3305|M.marinum_M LAAVRIALEDLDAQVSRLEDEIDAVRKREDRDRSLLTSGAVDAKQLADLQ
MUL_1323|M.ulcerans_Agy99 LGAVRIALEDLDAQVSRLEDEIDAVRKREDRDRSLLTSGAVDAKQLADLQ
Mvan_3619|M.vanbaalenii_PYR-1 LAALQLALADIDAQVAKLESEIDGVRQREDRDRALLDGGTVDPKQLTDLQ
Mflv_2891|M.gilvum_PYR-GCK VASLRLALADIDAQVAKFETEIDGVRQREDRDKALLEGGTVDPKQLTDLQ
MSMEG_4306|M.smegmatis_MC2_155 LAALGIALEDLDAQVAKYESEIDSVRQREDRDRALLEGGSVGAKQVTEIQ
TH_1466|M.thermoresistible__bu LAVLGIAVEDIDAEAAKLESEIEAVRRREERDRAMLEGGGVNAKQLTELQ
MAB_1904|M.abscessus_ATCC_1997 VGALAIALEDLDEQVAKLDAEVTAVRQREDRDRSLLASGSTNAKELTEIQ
MAV_2210|M.avium_104 LGAVRIALEDIDAHVRRLEAEVDAVRQREDRDRSLLQSGAIDAKQLADLQ
MLBr_01638|M.leprae_Br4923 LVAVQIALEDVDTQVFLLESEIDAMRQREDRDRLLLNSGATDAKQLSDLQ
: : :* *:* .. : *: .:*:* :**: :* .* ..*:::::*
Mb2254c|M.bovis_AF2122/97 HELDSLQRRQASLEDALLEVLERREELQAQQTAESRALQALRADLAAAQQ
Rv2229c|M.tuberculosis_H37Rv HELDSLQRRQASLEDALLEVLERREELQAQQTAESRALQALRADLAAAQQ
MMAR_3305|M.marinum_M HELETLERRQASLEDSLLEVMERREELQAQQNTEIAALEVLQAELTAAQQ
MUL_1323|M.ulcerans_Agy99 HELETLERRQASLEDSLLEVMERREELQAQQNTEIAALEVLQAELTAAQQ
Mvan_3619|M.vanbaalenii_PYR-1 HELDTLQRRQASLEDSQLEVMERREELAAEQTREQAAIEELQTALTGAQR
Mflv_2891|M.gilvum_PYR-GCK HELETLQRRQASLEDSQLELMERREELATREAEEASAAGQAQTALDDAQR
MSMEG_4306|M.smegmatis_MC2_155 HELETLQRRQASLEEQLLEVMERREELMAERSEELRRVDELQTELTEAQQ
TH_1466|M.thermoresistible__bu HELQTLERRKSSLEDSLLDVMERREQLDAERAGVSAEVDELQQQVQNAQQ
MAB_1904|M.abscessus_ATCC_1997 HELDTLERRQSSLEDSELELMERREELQKQQASAQAKADTIAERLSEIER
MAV_2210|M.avium_104 HELETLQRRQTSLEDSLLEVMERREELQAQLDGEQQTLKELEAAMATARR
MLBr_01638|M.leprae_Br4923 PEFGTWQRRKNSLEDSLREVMKRRGELQDQLTAELGAIERMQTDLVGARQ
*: : :**: ***: ::::** :* . : .:
Mb2254c|M.bovis_AF2122/97 ALDEALAEIDQARHQHSSQRDMLTATLDPELAGLYERQRAGGGPGAGRLQ
Rv2229c|M.tuberculosis_H37Rv ALDEALAEIDQARHQHSSQRDMLTATLDPELAGLYERQRAGGGPGAGRLQ
MMAR_3305|M.marinum_M SVDAALAELDQSRQEHSSRRDTLAASLNPDLAALYERLRAGGGPGAGQLQ
MUL_1323|M.ulcerans_Agy99 SVDAALAELDQSRQEHSSRRDTLAASLNPDLAALYERLRAGGGPGAGQLQ
Mvan_3619|M.vanbaalenii_PYR-1 ACDDARAELAQAREKSAARRAELVSAIDGELVALYERQRARGGVGAGVLQ
Mflv_2891|M.gilvum_PYR-GCK VCDDARAELVQTRERATARRAELADTIDGELVSLYERQRSRSGVGAAPLQ
MSMEG_4306|M.smegmatis_MC2_155 ARDAALVELDQARHQCATRRDALVNAIDDQLVELYEKQRARGGAGAGPLQ
TH_1466|M.thermoresistible__bu TVDDALAELARTRQQRTARREELIAELDAELVALYERQRTRGGVGAGPLQ
MAB_1904|M.abscessus_ATCC_1997 IQRAVAIDTDAEENQVRQRRDGLASSIDGLLLETYERQRRSGGAGAGFLQ
MAV_2210|M.avium_104 DLDAARGEISESRALHSSRREALSAELDPELFALYERQRARGGPGAGQLL
MLBr_01638|M.leprae_Br4923 TLDVAFAEIDQVGQPHSSQCDVLIAELAPALSAPYERLCAGGGLGVGQLQ
. : : * : * **: .* *.. *
Mb2254c|M.bovis_AF2122/97 GHRCGACRIEIGRGELAQISAAAEDEVVRCPECGAILLQLEGFEE
Rv2229c|M.tuberculosis_H37Rv GHRCGACRIEIGRGELAQISAAAEDEVVRCPECGAILLRLEGFEE
MMAR_3305|M.marinum_M GHRCGACRIEIGRSELARISAAAEDEVVRCPECAAILLRIKGPEL
MUL_1323|M.ulcerans_Agy99 GHRCGACRIEIGRSELARISAAAEDEVVRCPECAAILLRIKGPEL
Mvan_3619|M.vanbaalenii_PYR-1 GRRCGACRIEIDQGESARIAAAAEDEVLRCPECSAILLRVKGSRR
Mflv_2891|M.gilvum_PYR-GCK GRRCGACRIEIDRGESARIAAAADDDVVRCPECSAILLRVKASRA
MSMEG_4306|M.smegmatis_MC2_155 GRRCGACRIEIDRGEIARITAAADDDVVRCPECGAILLRVKQ---
TH_1466|M.thermoresistible__bu GGRCGACQIEIDRGELARFSAAAEDDVLRCPECGAILLRVKEFNR
MAB_1904|M.abscessus_ATCC_1997 GNKCGACRIELDRGELARISAADADEVLRCPECSAILVRP-----
MAV_2210|M.avium_104 GRRCGACRLEIDRGELSRISAAAEDDVVRCPECGAILLRVKGPGQ
MLBr_01638|M.leprae_Br4923 GHRCGACRSEIGRGELSCISVDVDDEVVKYPESGAIQLLDKGFFQ
* :****: *:.:.* : ::. *:*:: **..** :