For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTTPAPPQAPRTDLSDHLRRLREFTLSPQARPALVGALGATLITAGGLGAGSTRLHDPVLESMHLSWLRF GHGLVVSSLLLWGGVALMLMAWLWLGRRVVDRTATEYTMIATTGFWLAPLLLSVPVFSRDTYSYLAQGAL LRDGFDPYVVGPVENPNSLLDDVSPIWTTTTAPYGPAFILVAKFVTILVGNDVVAGTMLLRLCMLPGLAL LIWAVPRVARHVGASGVAALWIGVLNPLMIVHLMGGVHNEMLMVGLMMAGIALCLSGRNVWGITLIAVAV AVKATAGLALPFMVWVWARRLRDRRGLAPVKAFAAATAASLVIFVAVFSVLSLAAGVGLGWLNALAAGNV KIINWLTVPTALANLINAVGGLVFPVNFYAVIDVTRIVGLAIIAISAPVLWWRLRHTDREALMGIALVMA VVVLFVPAALPWYYTWPLAVVAALATSRRSMALIAGFSTWITVIWRPDGAHGMYSWGHVLLSLSCAAAAW YWLYKAPEGTAEAVSTPSPARDEPPHEPDASATHRPGGR
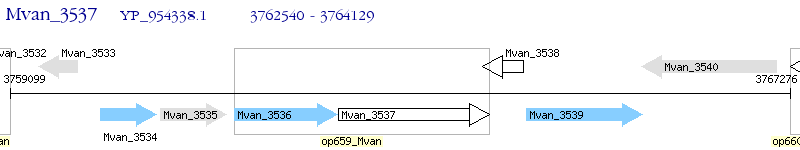
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_3537 | - | - | 100% (529) | hypothetical protein Mvan_3537 |
| M. vanbaalenii PYR-1 | Mvan_2731 | - | 6e-62 | 31.47% (537) | hypothetical protein Mvan_2731 |
| M. vanbaalenii PYR-1 | Mvan_4862 | - | 8e-06 | 29.68% (155) | glycosyl transferase family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2196 | - | 0.0 | 69.26% (514) | integral membrane protein |
| M. gilvum PYR-GCK | Mflv_2974 | - | 0.0 | 84.50% (516) | hypothetical protein Mflv_2974 |
| M. tuberculosis H37Rv | Rv2174 | - | 0.0 | 69.07% (514) | integral membrane protein |
| M. leprae Br4923 | MLBr_00899 | - | 0.0 | 68.13% (502) | putative integral-membrane protein |
| M. abscessus ATCC 19977 | MAB_1991c | - | 0.0 | 61.86% (506) | hypothetical protein MAB_1991c |
| M. marinum M | MMAR_3209 | - | 0.0 | 68.73% (518) | hypothetical protein MMAR_3209 |
| M. avium 104 | MAV_2319 | - | 0.0 | 70.22% (497) | integral membrane protein |
| M. smegmatis MC2 155 | MSMEG_4241 | - | 0.0 | 77.58% (504) | integral membrane protein |
| M. thermoresistible (build 8) | TH_0709 | - | 0.0 | 76.57% (495) | Possible conserved integral membrane protein |
| M. ulcerans Agy99 | MUL_3517 | - | 0.0 | 70.19% (473) | hypothetical protein MUL_3517 |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_3537|M.vanbaalenii_PYR-1 ---------MTTPAPPQAPRTDLSDHLRRLREFTLSPQARPALVGALGAT
Mflv_2974|M.gilvum_PYR-GCK ---------MTSSMSQTAAAP---NHLRRLKEFTLSRQGRPALVGALGAV
MSMEG_4241|M.smegmatis_MC2_155 ----------MTPTETHKPNPGLAEHVSRLAAFAVSAEARPARLGCLGAT
TH_0709|M.thermoresistible__bu --------MTASTSSTDLPKSGLGGQISRLNQFARSAAGRPALLGGLGAV
Mb2196|M.bovis_AF2122/97 ----MTTPSHAPAVDLATAKDAVVQHLSRLFEFTTGPQGGPARLGFAGAV
Rv2174|M.tuberculosis_H37Rv ----MTTPSHAPAVDLATAKDAVVQHLSRLFEFTTGPQGGPARLGFAGAV
MMAR_3209|M.marinum_M ----MTSPTPTPEA-LSVTKMSVRQRLSRIREFASSPQAGPARLGLLGAV
MUL_3517|M.ulcerans_Agy99 -------------------------------------------------M
MAV_2319|M.avium_104 ----MATGTDTPAADPPAAKPSRGQWFSRLKAFAGSADSGPAKLGMLGSV
MLBr_00899|M.leprae_Br4923 ----MTTPTHLPEANSPTKR-YIHQYFSQLRMFAASPESGPAKLGLLGSL
MAB_1991c|M.abscessus_ATCC_199 MSTSSKALEPSQEPKPDPDSPAPAPAPGWVSSFLRSPAGRAATIGFGGAI
Mvan_3537|M.vanbaalenii_PYR-1 LITAGGLGAGSTRLHDPVLESMHLSWLRFGHGLVVSSLLLWGGVALMLMA
Mflv_2974|M.gilvum_PYR-GCK LLTAGGLGAGSTRLHDPLLESMHLSWLRFGHGLVVSSLLLWGGVALMVLA
MSMEG_4241|M.smegmatis_MC2_155 VLTIGGLGAGSTRLHDPLLESLHLSWLRFGHGLVVSSILLWTGVALMLIA
TH_0709|M.thermoresistible__bu LITAGGLGAGSTRVHDPLLESLYLSWLRFGHGLVVSSTLLWTGVTLMLIA
Mb2196|M.bovis_AF2122/97 LITAGGLGAGSVRQHDPLLESIHMSWLRFGHGLVLSSILLWTGVGVMLLA
Rv2174|M.tuberculosis_H37Rv LITAGGLGAGSVRQHDPLLESIHMSWLRFGHGLVLSSILLWTGVGVMLLA
MMAR_3209|M.marinum_M LITVGGLGAGSIRQHDPLLESIHMSWLRFGHGLVLSSIVLWLGVGLMLIA
MUL_3517|M.ulcerans_Agy99 LITVGGLGAGSIRQHDPLLESIHMSWLRFGHGLVLSSIVLWLGVGLMLIA
MAV_2319|M.avium_104 LITLGGLGAGSTRQHDPLLESIHMSWLRFGHGLVLSSIVLWTGVGLMLIA
MLBr_00899|M.leprae_Br4923 LITTGGLGAGSIRQHDPLLELICMSWLRFGHGLVLSSILLWTGVSLMLIA
MAB_1991c|M.abscessus_ATCC_199 LVAIGGLGAGSTRQHDPLLESAGLSWLRFGHGLVVSSICMWLGVVLMLLA
::: ******* * ***:** :**********:** :* ** :*::*
Mvan_3537|M.vanbaalenii_PYR-1 WLWLGRRVVDRTATEYTMIATTGFWLAPLLLSVPVFSRDTYSYLAQGALL
Mflv_2974|M.gilvum_PYR-GCK WVWLGRHVIDRTATQYTMLATTAFWLTPLLLSVPLFSRDTYSYLAQGALL
MSMEG_4241|M.smegmatis_MC2_155 WLWLGRRVIDRTATEYTMVATTGFWLAPLLLSVPLFSRDTYSYLAQGALL
TH_0709|M.thermoresistible__bu WLWLGRHVITGRATEYGMIATTGFWLVPLLLSVPVFSRDTYSYLAQGALL
Mb2196|M.bovis_AF2122/97 WLGLGRRVLAGEATEFTMRATTVIWLAPLLLSVPVFSRDTYSYLAQGALL
Rv2174|M.tuberculosis_H37Rv WLGLGRRVLAGEATEFTMRATTVIWLAPLLLSVPVFSRDTYSYLAQGALL
MMAR_3209|M.marinum_M WLRLGRGVLAGQASEFTMRATTAFWLAPLLLSVPIFSRDTYSYLAQGALL
MUL_3517|M.ulcerans_Agy99 WLRLGRGVLAGQASEFTMRATTAFWLAPLLLSVPIFSRDTYSYLAQGALL
MAV_2319|M.avium_104 WLSLGRRVLAGEATEFVMKATTGFWLAPLLVSVPVFSRDTYSYLAQGALL
MLBr_00899|M.leprae_Br4923 WLRLGRRVLAGQATEFMMRATIGFWLTPLLLSVPVFSRDTYSYLAQGALL
MAB_1991c|M.abscessus_ATCC_199 WLGLGRHVIAGKVTKESLLIVIPCWLLPLLASVPVFSRDAYSYLAQGALL
*: *** *: .:: : . ** *** ***:****:**********
Mvan_3537|M.vanbaalenii_PYR-1 RDGFDPYVVGPVENPNSLLDDVSPIWTTTTAPYGPAFILVAKFVTILVGN
Mflv_2974|M.gilvum_PYR-GCK RDGFDPYVVGPIENPNSLLDNVSPIWTTTTAPYGPAFILVAKFVTMLVGD
MSMEG_4241|M.smegmatis_MC2_155 RDGFDPYVVGPVENPNSLLENVSPIWTTTTAPYGPAFILVAKFVTMLVGN
TH_0709|M.thermoresistible__bu RDGFDPYLVGPVENPNPLLDDVSPIWTTTTAPYGPAFILVAKFVTMLVND
Mb2196|M.bovis_AF2122/97 RDGLDPYAVGPVGNPNALLDDVSPIWTITTAPYGPAFILVAKFVTVIVGN
Rv2174|M.tuberculosis_H37Rv RDGLDPYAVGPVGNPNALLDDVSPIWTITTAPYGPAFILVAKFVTVIVGN
MMAR_3209|M.marinum_M RDGLDPYAVGPVANANSLLNDVSPIWTITTAPYGPAFILVAKVVTMIVGN
MUL_3517|M.ulcerans_Agy99 RDGLDPYAVSPVANANSLLNDVSPIWTITTAPYGPAFILVAKVVTMIVGN
MAV_2319|M.avium_104 RDGLDPYAVGPVANPNSLLDNVSPIWSITTAPYGPVFILVAKIITILVGN
MLBr_00899|M.leprae_Br4923 RDGLDPYLVGPVGNPNALLDNVSPIWTITTAPYGPAFILVAKFVTLAVGN
MAB_1991c|M.abscessus_ATCC_199 RDGFDPYLVGPVENPNALLENVSPIWTTTTAPYGPLFLLIARFVTQIVGD
***:*** *.*: *.*.**::*****: ******* *:*:*:.:* *.:
Mvan_3537|M.vanbaalenii_PYR-1 DVVAGTMLLRLCMLPGLALLIWAVPRVARHVGASGVAALWIGVLNPLMIV
Mflv_2974|M.gilvum_PYR-GCK DVVAGTMLLRLCMLPGLALLIWAAPRVARHVGASGAAALWIAVLNPLVII
MSMEG_4241|M.smegmatis_MC2_155 DVVAGTMLLRLCMLPGLLLLIWAAPRVARHVGANAAAALWICVLNPLVII
TH_0709|M.thermoresistible__bu NVVAGTMLLRLCMLPGLALLIWAAPRVARHIGANPAAALWICVLNPLVII
Mb2196|M.bovis_AF2122/97 NVVAGTMLLRLCMLPGLALLVWATPRLASHLGTHGPTALWICVLNPLVLI
Rv2174|M.tuberculosis_H37Rv NVVAGTMLLRLCMLPGLALLVWATPRLASHLGTHGPTALWICVLNPLVLI
MMAR_3209|M.marinum_M NVVAGTMLLRLCMLPGLALLIWATPRLAHHLGADSPTAMWICVLNPLVLI
MUL_3517|M.ulcerans_Agy99 NVVAGTMLLRLCMLPGLALLIWATPRLAHHLGADSPTAMWICVLNPLVLI
MAV_2319|M.avium_104 NVVAGTALLRLCMLPGLVLLIWATPRLARHLGADEPTALWICVLNPLVLI
MLBr_00899|M.leprae_Br4923 NVVTGTMLLRLCMLPGLALLIWATPRLAQHFGADGTTALWTCVLNPLVLI
MAB_1991c|M.abscessus_ATCC_199 DVVAGTMLLRLCMLPGLALLVWATPRVAQFFSASASKALWICVLNPLVII
:**:** ********** **:**.**:* ...: *:* *****:::
Mvan_3537|M.vanbaalenii_PYR-1 HLMGGVHNEMLMVGLMMAGIALCLSGRNVWGITLIAVAVAVKATAGLALP
Mflv_2974|M.gilvum_PYR-GCK HLMGGVHNEMLMVGLMMAGIALCFAGRNVSGVTLIAVAVAVKATAGLALP
MSMEG_4241|M.smegmatis_MC2_155 HLMGGVHNEMLMVGLMMAAIALTFAGRPIWGVGLIATAVAVKATAGLALP
TH_0709|M.thermoresistible__bu HLMGGVHNEMLMVGLMMAGIALTLSRHPVWGVASIAVAVAVKATAGLALP
Mb2196|M.bovis_AF2122/97 HLMGGVHNEMLMVGLMTAGIALTVQGRNVAGIILITVAIAVKATAGIALP
Rv2174|M.tuberculosis_H37Rv HLMGGVHNEMLMVGLMTAGIALTVQGRNVAGIILITVAIAVKATAGIALP
MMAR_3209|M.marinum_M HLMGGVHNEMLMVGLMAAGIALTLQRHHVAGIALITVAVAVKATAGLALP
MUL_3517|M.ulcerans_Agy99 HLMGGVHNEMLMVGLMAAGIALTLQRHHVAGIALITVAVAVKATAGLALP
MAV_2319|M.avium_104 HLMGGVHNEMLMVGLMAAGIALTFGGRPVAGVTLITVAIAVKATAGIALP
MLBr_00899|M.leprae_Br4923 HLMGGVHNEMLMVGLMTAGVALTFQGRNVAGTALITVAIAVKATAGISLP
MAB_1991c|M.abscessus_ATCC_199 HLMGGVHNEMLMVGLMMAGIALTLERHHVWGIAVVALGVMVKASAGLALP
**************** *.:** . : : * :: .: ***:**::**
Mvan_3537|M.vanbaalenii_PYR-1 FMVWVWARRLRDRRG-----LAPVKAFAAATAASLVIFVAVFSVLSLAAG
Mflv_2974|M.gilvum_PYR-GCK FMVWVWARQLRDRRG-----FSPAKAFAAATVGSVLVFVVVFAVLSLAAG
MSMEG_4241|M.smegmatis_MC2_155 FMVWVWMRQLRDRHD-----MPPVRAFVVSTAASVLIFGAVFAVLSWVAG
TH_0709|M.thermoresistible__bu FMVWVWMRQLREQRS-----LSPAAAFAIATAASLAIFAAVFAVLSWVAG
Mb2196|M.bovis_AF2122/97 FLVWVWLRHLRERRG-----YRPVQAFLAAAAISLLIFVAVFAVLSAVAG
Rv2174|M.tuberculosis_H37Rv FLVWVWLRHLRERRG-----YRPVQAFLAAAAISLLIFVAVFAVLSAVAG
MMAR_3209|M.marinum_M FLFWVWMRHLHERRG-----YRPAHAFLVAAAASLLVFLAVFSVLSAVAR
MUL_3517|M.ulcerans_Agy99 FLFWVWMRHLHERRG-----YRPAHAFLVAAAASLLVFLAVFSVLSAVAR
MAV_2319|M.avium_104 FLVWVWARQLRDQRG-----YRPVPAFFAATAASLLIFAVVFAALSAVAG
MLBr_00899|M.leprae_Br4923 FMVWVWMRQLHNRRG-----YRPVQAFLAATATALLIFATVFAVLSTAAS
MAB_1991c|M.abscessus_ATCC_199 FLVWIWMRRLASDHVDGAPPRRPPAAFALASIGSVAISLTTFALMSWITG
*:.*:* *:* . : * ** :: :: : ..*: :* :
Mvan_3537|M.vanbaalenii_PYR-1 VGLGWLNALAAGNVKIINWLTVPTALANLINAVGGLVFPVNFYAVIDVTR
Mflv_2974|M.gilvum_PYR-GCK VGLGWLTALAG-SVKIINWLTVPTAIANLTNAVVGLVMPVNFYAVLETTR
MSMEG_4241|M.smegmatis_MC2_155 VGLGWLTALAG-SVKIINWLTVPTAASNLINAIGGLFFSVNFYAVLEVAR
TH_0709|M.thermoresistible__bu VGLGWLTALAG-SVKIINWLTVPTAVANLTNAIGGMFLPVNFYAVLEVTR
Mb2196|M.bovis_AF2122/97 VGLGWLTALAG-SVKIINWLTVPTGAANVIHALGRGLFTVDFYTLLRITR
Rv2174|M.tuberculosis_H37Rv VGLGWLTALAG-SVKIINWLTVPTGAANVIHALGRGLFTVDFYTLLRITR
MMAR_3209|M.marinum_M VGLGWLTALAG-SVKIINWLTLPTGAANLIHALGRNFFTLDFYTLLHVTR
MUL_3517|M.ulcerans_Agy99 VGLGWLTALAG-SVKIINWLTLPTGAANLIHALGRNFFTLDFYTLLHVTR
MAV_2319|M.avium_104 VGLGWLTALAG-SVKIINWLTVPTAAANLIHAIGSGFFPVSFYPILRVTR
MLBr_00899|M.leprae_Br4923 VGLGWMTALAG-SVKIINWLTVPTAIANLVNVLANGFFATDFYGVLRMTR
MAB_1991c|M.abscessus_ATCC_199 VGFGWLTAFAGSAVKIINWLTIPTAVANVINVVAGLFVTVNFDAVLEVMR
**:**:.*:*. ********:**. :*: :.: ... .* :: *
Mvan_3537|M.vanbaalenii_PYR-1 IVGLAIIAISAPVLWWRLRHTDREALMGIALVMAVVVLFVPAALPWYYTW
Mflv_2974|M.gilvum_PYR-GCK IIGIGVIAISAPVLWWRYRRTDRDALMGIALVMAVVVLFVPAALPWYYTW
MSMEG_4241|M.smegmatis_MC2_155 IIGIAAIVIALPLLWWRYRHTDREALTGIALAMVVVVLFVPAALPWYYTW
TH_0709|M.thermoresistible__bu IIGIAIIVVTLPVVWWRFRHDDRGALTGIAVAMLVVVLFVPAALPWYYTW
Mb2196|M.bovis_AF2122/97 LIGIVIIAVSLPLLWWRFRRDDRAALTGVAWSMLIVVLFVPAALPWYYSW
Rv2174|M.tuberculosis_H37Rv LIGIVIIAVSLPLLWWRFRRDDRAALTGVAWSMLIVVLFVPAALPWYYSW
MMAR_3209|M.marinum_M LIGIAIIAVSLPLLWWRFRRDDRAALTGIAWSMLIVVLFVPAALPWYYSW
MUL_3517|M.ulcerans_Agy99 LIGIAIIAVSLPLLWWRFRRDDRAALTGIAWSMLIVVLFVPAALPWYYSW
MAV_2319|M.avium_104 LVGIAIIAISLPLLWWRFRRDDRDALTGIAISMLIVVLFVPAALPWYYSW
MLBr_00899|M.leprae_Br4923 FIGIVIIAVSLPLLWWRYRRDDRSALTGIAWSMLIVVLFVPAALPWYYSW
MAB_1991c|M.abscessus_ATCC_199 IIGVLVIAVSLPVVWWRHRHNERDAMFGILWAMSIVVLMAPAALPWYYSW
::*: *.:: *::*** *: :* *: *: * :***:.********:*
Mvan_3537|M.vanbaalenii_PYR-1 PLAVVAALATSRRSMALIAGFSTWITVIWRPDGAHGMYSWGHVLLSLSCA
Mflv_2974|M.gilvum_PYR-GCK PLAVIAALAQSRRSIALIAGFSTWITAIWRPDGAHGMYSWAHVLLATACA
MSMEG_4241|M.smegmatis_MC2_155 PLAVAAALVQTRPGIAAIAGFSTWIMVMWKPDGAHGMYSWAHVLLTTACA
TH_0709|M.thermoresistible__bu PLAVLSALAQSRPAIALIAGFSTWIMVIFRPDGAHGMYSWTHVLIATGCA
Mb2196|M.bovis_AF2122/97 PLAVAAPLAQSRRAIAAIAGLSTWVMVIFKPDGSHGMYSWLHFWIATACA
Rv2174|M.tuberculosis_H37Rv PLAVAAPLAQARRAIAAIAGLSTWVMVIFKPDGSHGMYSWLHFWIATACA
MMAR_3209|M.marinum_M PLAVAAPLAQSRRSMAAIAGISTWIMVIFKPDGSHGMYSWLHFWIATSCA
MUL_3517|M.ulcerans_Agy99 PLAVAAPLAQSRRSMAAVAGISTWIMVIFKPDGSHGMYSWLHFWIATGCA
MAV_2319|M.avium_104 PLAVLAPLAQSRRAVAIIGGLSTWVMVIFKPDGSHGMYSWLHFSLATAVA
MLBr_00899|M.leprae_Br4923 PLAVAAPLAQSRQALATIAGFSSWIMVIFKPDGSHGMYSWLHFSLATTCA
MAB_1991c|M.abscessus_ATCC_199 PLAIAAPLLQSRGAIAAIAGFSTWIMVIFKPDGSHGMYVWIHVLIAATCA
***: :.* :* .:* :.*:*:*: .:::***:**** * *. :: *
Mvan_3537|M.vanbaalenii_PYR-1 AAAWYWLYKAPEGT--AEAVSTPSPARDEPPHEPDASATHRPGGR-----
Mflv_2974|M.gilvum_PYR-GCK AAAWYWLSKQPEGADRPEDFSTPSPARDEPPRGPDASAARRPDAR-----
MSMEG_4241|M.smegmatis_MC2_155 VAAWYSLYRAPESA--------------------EATASRQ---------
TH_0709|M.thermoresistible__bu VAAWWWLYRTESTP---------------AEPIPQSGGSAGESGR-----
Mb2196|M.bovis_AF2122/97 LTAWYVLYRSPDRR--------------GVQAATPVVNTP----------
Rv2174|M.tuberculosis_H37Rv LTAWYVLYRSPDRR--------------GVQAATPVVNTP----------
MMAR_3209|M.marinum_M LAAWYALYRAPDLR--------------TRPGETPADEPAAPLDMGTKDP
MUL_3517|M.ulcerans_Agy99 LAAWYALYRAPDLR--------------TRPGETPADEPAAPLDMGTKDP
MAV_2319|M.avium_104 LVAWYSLYRVP----------------------QPAQ-------------
MLBr_00899|M.leprae_Br4923 LIAWHALYKAP---------------------ETDTA-------------
MAB_1991c|M.abscessus_ATCC_199 AIAWRMINTAPE---------------------PDGSAPAA---------
** :
Mvan_3537|M.vanbaalenii_PYR-1 -
Mflv_2974|M.gilvum_PYR-GCK -
MSMEG_4241|M.smegmatis_MC2_155 -
TH_0709|M.thermoresistible__bu -
Mb2196|M.bovis_AF2122/97 -
Rv2174|M.tuberculosis_H37Rv -
MMAR_3209|M.marinum_M Q
MUL_3517|M.ulcerans_Agy99 H
MAV_2319|M.avium_104 -
MLBr_00899|M.leprae_Br4923 -
MAB_1991c|M.abscessus_ATCC_199 -