For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MKTRVPRHFVAALIGGGAVLGVAGCSTPEPALGGTTATVTIDGSDTGGAHAVRCRQNGWSWYIDTTEKDN GFTAVLSTGGDVTAESVDFRGVGGFTGSFWADNIGDAEVTGRDGSYTITGKADGTFAGKPSEAVTAEFRI QANC
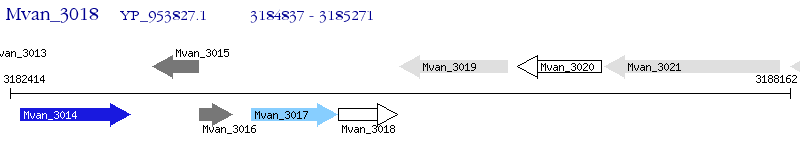
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_3018 | - | - | 100% (144) | hypothetical protein Mvan_3018 |
| M. vanbaalenii PYR-1 | Mvan_5581 | - | 1e-15 | 35.82% (134) | hypothetical protein Mvan_5581 |
| M. vanbaalenii PYR-1 | Mvan_5582 | - | 4e-10 | 28.57% (147) | hypothetical protein Mvan_5582 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1913c | lppE | 2e-11 | 32.80% (125) | lipoprotein LppE |
| M. gilvum PYR-GCK | Mflv_3293 | - | 1e-56 | 71.01% (138) | hypothetical protein Mflv_3293 |
| M. tuberculosis H37Rv | Rv1881c | lppE | 2e-11 | 32.80% (125) | lipoprotein LppE |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2379 | - | 1e-07 | 28.10% (153) | lipoprotein LpqH precursor |
| M. marinum M | MMAR_4690 | - | 3e-17 | 36.72% (128) | lipoprotein |
| M. avium 104 | MAV_2821 | - | 2e-17 | 38.35% (133) | LppE protein |
| M. smegmatis MC2 155 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_3147 | - | 4e-31 | 47.54% (122) | PUTATIVE putative conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_0309 | - | 2e-16 | 35.94% (128) | lipoprotein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1913c|M.bovis_AF2122/97 --MCN---RLVTVTGVAMVVAAGLSACG-------------------QAQ
Rv1881c|M.tuberculosis_H37Rv --MCN---RLVTVTGVAMVVAAGLSACG-------------------QAQ
MAV_2821|M.avium_104 --MQK---RLPTVVLVLTAVVAGIAGCS-------------------AAQ
Mvan_3018|M.vanbaalenii_PYR-1 --MKTRVPRHFVAALIGGGAVLGVAGCST------------------PEP
Mflv_3293|M.gilvum_PYR-GCK --MLT---RHLMVAVAGSALVLGVAGCST------------------PEP
TH_3147|M.thermoresistible__bu VGIRVAPNRGPWTLALLAAVGL-AAGCSA------------------APS
MMAR_4690|M.marinum_M --MRN----LIKLAFVCVLASAGLTGCGS------------------SSG
MUL_0309|M.ulcerans_Agy99 --MRN----PIKLAFVCLLASAGLTGCGS------------------SSG
MAB_2379|M.abscessus_ATCC_1997 --MQR--TGSVVVIGAGAIIALGLAGCSGDGNESAPSSAEKTTVSAEGGA
: :.*.
Mb1913c|M.bovis_AF2122/97 TVPRKAARLTIDG-VTHTTRPATCSQEHSYRTIDIRNHDSTVQAVVLLSG
Rv1881c|M.tuberculosis_H37Rv TVPRKAARLTIDG-VTHTTRPATCSQEHSYRTIDIRNHDSTVQAVVLLSG
MAV_2821|M.avium_104 TVPRKAARLTIDG-ATHTTRPTSCRQDQMYRTINIPDHDGGVEAVVLLSG
Mvan_3018|M.vanbaalenii_PYR-1 ALGGTTATVTIDGSDTGGAHAVRCRQNGWSWYIDTTEKDNGFTAVLSTGG
Mflv_3293|M.gilvum_PYR-GCK ALGGTTATVSIDGKDTGGAHVVRCHQSGWSWFIETPDEENGFVAVVSTGE
TH_3147|M.thermoresistible__bu ALGDSTAEVKINGEAAGGPYPVTCSQNGWAWTVQTQQQDTGFTAVFDTEP
MMAR_4690|M.marinum_M SQLASTASMTVNGADVH-PRVVRCTQIEWYRTIQIGDQASGVRVLIDQGA
MUL_0309|M.ulcerans_Agy99 SQLASTASMTVNGADVQ-PRVVRCTQIEWYRTIQIGDQASGVRILIDQGA
MAB_2379|M.abscessus_ATCC_1997 AAMNSSAKVVVDGKEKKVEGAIACSTAAGQVNIGMGAGMQAIAVVLSDGD
: .:* : ::* * : . :.
Mb1913c|M.bovis_AF2122/97 DRVIPQWVKIRNVDGFNGSFWHGGVGNARADRARNT-YTVAGSAYGISSK
Rv1881c|M.tuberculosis_H37Rv DRVIPQWVKIRNVDGFNGSFWHGGVGNARADRARNT-YTVAGSAYGISSK
MAV_2821|M.avium_104 YRVMPQWVKIRNVDGFTGSYWQGGVGDAHVDLTNDA-YTITGSAYGINSA
Mvan_3018|M.vanbaalenii_PYR-1 -DVTAESVDFRGVGGFTGSFWADNIGDAEVTGRDGS-YTITGKADGTFAG
Mflv_3293|M.gilvum_PYR-GCK -KVTAESVNFRGFGGFTGSFWADNIGDAEVSSDGGS-FTISGTADGAFDD
TH_3147|M.thermoresistible__bu -EMTAQSVEITGLNGFTGSFWAGTVGKGRAGVANGK-FRISGTAVGSFAD
MMAR_4690|M.marinum_M RPMIAKSVRITNLGGFTGMYAQGDGGDAGLHFSDGR-FTITGNADGYNTD
MUL_0309|M.ulcerans_Agy99 RPMIAKSVRITNLGGFTGMYAQGDGGDAGLHFSDGR-FTITRNADGYNTD
MAB_2379|M.abscessus_ATCC_1997 -SPKVTSVGLGNIDGVTLGFADGAPGGKATATKDGKKYTVTGTATGVDTA
* : ...*.. : . * . : :: .* *
Mb1913c|M.bovis_AF2122/97 KPNTVVSTDFNILAEC-
Rv1881c|M.tuberculosis_H37Rv KPNTVVSTDFNILAEC-
MAV_2821|M.avium_104 NPNKVVTTDFKIVAEC-
Mvan_3018|M.vanbaalenii_PYR-1 KPSEAVTAEFRIQANC-
Mflv_3293|M.gilvum_PYR-GCK TPSEAVSAKFRIEADC-
TH_3147|M.thermoresistible__bu NPTDTVDAEFSIEARC-
MMAR_4690|M.marinum_M KPAEPTTAAFRISVNC-
MUL_0309|M.ulcerans_Agy99 KPAEPTTAAFRISVNC-
MAB_2379|M.abscessus_ATCC_1997 NPMQPIEKPFEIEVTCP
.* * * . *