For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MLTRHLMVAVAGSALVLGVAGCSTPEPALGGTTATVSIDGKDTGGAHVVRCHQSGWSWFIETPDEENGFV AVVSTGEKVTAESVNFRGFGGFTGSFWADNIGDAEVSSDGGSFTISGTADGAFDDTPSEAVSAKFRIEAD C
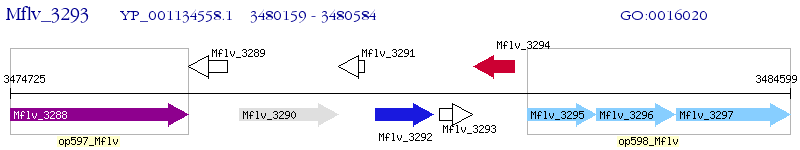
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3293 | - | - | 100% (141) | hypothetical protein Mflv_3293 |
| M. gilvum PYR-GCK | Mflv_3807 | - | 1e-20 | 40.74% (135) | hypothetical protein Mflv_3807 |
| M. gilvum PYR-GCK | Mflv_1227 | - | 7e-16 | 32.41% (145) | hypothetical protein Mflv_1227 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3789 | lpqH | 8e-12 | 33.33% (156) | 19 kDa lipoprotein antigen precursor LPQH |
| M. tuberculosis H37Rv | Rv3763 | lpqH | 9e-12 | 33.33% (156) | 19 kDa lipoprotein antigen precursor LPQH |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0885c | - | 3e-09 | 29.94% (157) | lipoprotein lpqH precursor |
| M. marinum M | MMAR_4690 | - | 1e-16 | 34.75% (141) | lipoprotein |
| M. avium 104 | MAV_0969 | - | 7e-19 | 38.35% (133) | hypothetical protein MAV_0969 |
| M. smegmatis MC2 155 | MSMEG_6315 | - | 2e-05 | 26.79% (168) | lipoprotein LpqH |
| M. thermoresistible (build 8) | TH_3147 | - | 1e-30 | 47.54% (122) | PUTATIVE putative conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_0309 | - | 1e-15 | 34.40% (125) | lipoprotein |
| M. vanbaalenii PYR-1 | Mvan_3018 | - | 1e-56 | 71.01% (138) | hypothetical protein Mvan_3018 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3293|M.gilvum_PYR-GCK --MLT---RHLMVAVAGSALVLGVAGCST-----------------PEPA
Mvan_3018|M.vanbaalenii_PYR-1 --MKTRVPRHFVAALIGGGAVLGVAGCST-----------------PEPA
TH_3147|M.thermoresistible__bu VGIRVAPNRGPWTLALLAAVGL-AAGCSA-----------------APSA
MMAR_4690|M.marinum_M ------MRNLIKLAFVCVLASAGLTGCGS-----------------SSGS
MUL_0309|M.ulcerans_Agy99 ------MRNPIKLAFVCLLASAGLTGCGS-----------------SSGS
MAV_0969|M.avium_104 ------MRNRF-VAAGAALTVALLGACTS-----------------RPPA
Mb3789|M.bovis_AF2122/97 -----MKRGLTVAVAGAAILVAGLSGCSS-NKSTTGSGETTTAAGTTASP
Rv3763|M.tuberculosis_H37Rv -----MKRGLTVAVAGAAILVAGLSGCSS-NKSTTGSGETTTAAGTTASP
MAB_0885c|M.abscessus_ATCC_199 -----MKRGLVVAIGGAAIVAAGLVGCSG-DKDKSET-KGTTVSNSAGAV
MSMEG_6315|M.smegmatis_MC2_155 ---MELVVNRLVAAAGCAALLTGVAGCSSQDSNDAGQSPATESDGPAATP
.*
Mflv_3293|M.gilvum_PYR-GCK LGGTTATVSIDGKDTGGAHVVRCHQSGWSWFIETPDEENGFVAVVST-GE
Mvan_3018|M.vanbaalenii_PYR-1 LGGTTATVTIDGSDTGGAHAVRCRQNGWSWYIDTTEKDNGFTAVLST-GG
TH_3147|M.thermoresistible__bu LGDSTAEVKINGEAAGGPYPVTCSQNGWAWTVQTQQQDTGFTAVFDT-EP
MMAR_4690|M.marinum_M QLASTASMTVNGADVH-PRVVRCTQIEWYRTIQIGDQASGVRVLIDQGAR
MUL_0309|M.ulcerans_Agy99 QLASTASMTVNGADVQ-PRVVRCTQIEWYRTIQIGDQASGVRILIDQGAR
MAV_0969|M.avium_104 QLSSTASVTVDGKDRN-FHIVTCRQLEWRRMIDIGADFSGAKVAVDENAQ
Mb3789|M.bovis_AF2122/97 GAASGPKVVIDGKDQNVTGSVVCTTAAGNVNIAIGGAATGIAAVLTD-GN
Rv3763|M.tuberculosis_H37Rv GAASGPKVVIDGKDQNVTGSVVCTTAAGNVNIAIGGAATGIAAVLTD-GN
MAB_0885c|M.abscessus_ATCC_199 ATNSSAKVTIDGKEQKIEGAVVCQSSNGNVNIVIGSGAQGFGAVVTE-GD
MSMEG_6315|M.smegmatis_MC2_155 VEADQGKVVLGGNELGPVTAIGCETSDGLTTITIESDVR-TTVKVTE-GE
: :.* : * : .
Mflv_3293|M.gilvum_PYR-GCK KVTAESVNFRGFG--GFTGSFWADNIGDAEVSSDGG-SFTISGTADGAFD
Mvan_3018|M.vanbaalenii_PYR-1 DVTAESVDFRGVG--GFTGSFWADNIGDAEVTGRDG-SYTITGKADGTFA
TH_3147|M.thermoresistible__bu EMTAQSVEITGLN--GFTGSFWAGTVGKGRAGVANG-KFRISGTAVGSFA
MMAR_4690|M.marinum_M PMIAKSVRITNLG--GFTGMYAQGDGGDAGLHFSDG-RFTITGNADGYNT
MUL_0309|M.ulcerans_Agy99 PMIAKSVRITNLG--GFTGMYAQGDGGDAGLHFSDG-RFTITRNADGYNT
MAV_0969|M.avium_104 PPVVESVHIQNLS--GFSGMYSRGGSGSADMSMTGD-KFTISGTADGYKT
Mb3789|M.bovis_AF2122/97 PPEVKSVGLGNVN--GVTLGYTSGTGQGNASATKDGSHYKITGTATGVDM
Rv3763|M.tuberculosis_H37Rv PPEVKSVGLGNVN--GVTLGYTSGTGQGNASATKDGSHYKITGTATGVDM
MAB_0885c|M.abscessus_ATCC_199 SPTVKSAGLGSLN--GQSLAFSEGTPGASAKATKDGKKYHIEGTAIAVDM
MSMEG_6315|M.smegmatis_MC2_155 APNVESVNIGDVWSDGPSVVYMSGLSEVPVVATRDGDTYTVAGIGMGADP
.:*. : .. * : : . .. : : . .
Mflv_3293|M.gilvum_PYR-GCK DTPSEAVSAKFRIEADC-
Mvan_3018|M.vanbaalenii_PYR-1 GKPSEAVTAEFRIQANC-
TH_3147|M.thermoresistible__bu DNPTDTVDAEFSIEARC-
MMAR_4690|M.marinum_M DKPAEPTTAAFRISVNC-
MUL_0309|M.ulcerans_Agy99 DKPAEPTTAAFRISVNC-
MAV_0969|M.avium_104 DKPGEPATATFKIVVTC-
Mb3789|M.bovis_AF2122/97 ANPMSPVNKSFEIEVTCS
Rv3763|M.tuberculosis_H37Rv ANPMSPVNKSFEIEVTCS
MAB_0885c|M.abscessus_ATCC_199 ANPTQPSNKPFEIEVTCP
MSMEG_6315|M.smegmatis_MC2_155 ADPSVPADTPFDIAVTCP
* . * * . *