For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MWKCGRMRADRLVAILLLLQQRKQVTASEVALELEVSERTARRDLEALSGAGVPVYAMPGRGGGWRLLGG ARTDLSGLTASEARALFLVAGPASTATPSVKAALRKLVRALPEPFREQAEAAATSVVVDPRHWGSSQPEP RPPRFLDELQEAVIRGVQVRLGYVDRKGSQTERTVHPLGIVAKRPTWYLVSNTDAGRRTFRIDRVSSVEL TDDAVQRPQGFDLAESWREIADEVDRKRTPLEAQAMCAADGLGLLRMVLGGRLEVGGSTPDGRIEIVIRG HNEYALAGELAGLIEWLEVTGPQGVRDHLASIGGALVARYS
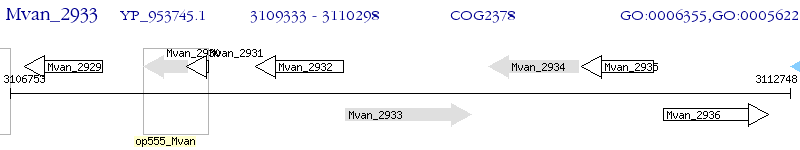
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_2933 | - | - | 100% (321) | helix-turn-helix, type 11 domain-containing protein |
| M. vanbaalenii PYR-1 | Mvan_1247 | - | 3e-17 | 31.71% (205) | helix-turn-helix, type 11 domain-containing protein |
| M. vanbaalenii PYR-1 | Mvan_2934 | - | 4e-15 | 31.16% (215) | helix-turn-helix, type 11 domain-containing protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_5106 | - | 4e-18 | 32.52% (206) | helix-turn-helix, type 11 domain-containing protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2185 | - | 7e-06 | 25.36% (343) | hypothetical protein MAB_2185 |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_2407 | - | 3e-06 | 28.14% (334) | hypothetical protein MAV_2407 |
| M. smegmatis MC2 155 | MSMEG_1359 | - | 4e-18 | 33.33% (210) | DeoR-family protein transcriptional regulator |
| M. thermoresistible (build 8) | TH_1858 | - | 7e-19 | 32.11% (218) | DeoR-family protein transcriptional regulator |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_1359|M.smegmatis_MC2_155 ---MARTVDTTGRVVQLLGLLQSRRVWTGEELAERLG---------VTGR
TH_1858|M.thermoresistible__bu MWQAADVSETTSRVLRLLGLLQSRRVWSGEELAERLG---------VTTR
Mflv_5106|M.gilvum_PYR-GCK ------MSETTGRVLQLLGLLQSRRVWSGDELAARLR---------VTTR
MAB_2185|M.abscessus_ATCC_1997 ----MAISKVERLMNLVICLLSTRTYVTAERIRTSVAGYSDSPSDEAFSR
MAV_2407|M.avium_104 --MPMATSKVERLVNLVIALLSTRGYITAEKIRSSVAGYSESASAEAFSR
Mvan_2933|M.vanbaalenii_PYR-1 --MWKCGRMRADRLVAILLLLQQRKQVTASEVALELE---------VSER
: :: **. * :...: : . *
MSMEG_1359|M.smegmatis_MC2_155 SVRRDIERLRELGYPVHASKGQ----GGGYQLG--AGMALPPLLLDPDEA
TH_1858|M.thermoresistible__bu SVRRDVERLRDLGYPVHASRGH----GGGYQLG--AGAALPPLLLDPEEA
Mflv_5106|M.gilvum_PYR-GCK SVRRDIDRLRELGYPVHASTGH----GGGYQLG--AGAALPPLLLDPDEA
MAB_2185|M.abscessus_ATCC_1997 MFERDKNELRDLGIPLETGKVSSFDATEGYRIRR-DAYELPDITLTADES
MAV_2407|M.avium_104 MFERDKNELRDLGIPLEVGRVSPMDPTEGYRINR-DAYALAPVDLTPDEA
Mvan_2933|M.vanbaalenii_PYR-1 TARRDLEALSGAGVPVYAMPGR----GGGWRLLGGARTDLSGLTASEARA
.** : * * *: . *::: *. : .:
MSMEG_1359|M.smegmatis_MC2_155 VAMAVCLRLAAGGSVAGVGESALRALSKLDQVMPARLRSQVAAIHDATVT
TH_1858|M.thermoresistible__bu VAVAVCLRLAAGGSIAGVGESALRALGKLDQVMPARLRSQVSAVHEATTT
Mflv_5106|M.gilvum_PYR-GCK VAVAVCLRVAAGGSVAGVGESALRALSKLDQVMPSRLRSQVAAVHDTTVT
MAB_2185|M.abscessus_ATCC_1997 AAVAVATQLWQSPELVTAAQGALMKLRAAGVDVDPAAEN--VSVAAPAAM
MAV_2407|M.avium_104 AAVAVATQLWESPELITATQGALLKLRAAGVDVDPLDDGAPVAIASPAGV
Mvan_2933|M.vanbaalenii_PYR-1 LFLVAGPASTATPSVKAALRKLVRALPEP---FREQAEAAATSVVVDPRH
:.. .: . . : * . :: .
MSMEG_1359|M.smegmatis_MC2_155 LGPNATDTAVAPDVLMTLARACRDREHVSTGYTDLRGN-QTQRRLEPYQL
TH_1858|M.thermoresistible__bu LGPNQPDQPVAPDVLMTLARACRDHEHVNTGYTDRAGN-RSRRRLEPYQL
Mflv_5106|M.gilvum_PYR-GCK LAG-ATDTPVEPEVLMTLARASRDHEHVTVGYTDVRGN-VTSRRVEPYQL
MAB_2185|M.abscessus_ATCC_1997 PGGRGSES-----VLRILLSAIDSGQAVQFGHRPSRTEPYVTRTVEPWGV
MAV_2407|M.avium_104 PGLRGSEE-----VLGILLSAIDSRQAVQFPHRPSRAEPYATRTVEPWGV
Mvan_2933|M.vanbaalenii_PYR-1 WGSSQPEPR-PPRFLDELQEAVIRGVQVRLGYVDRKGS-QTERTVHPLGI
. .: .* * * * : . * :.* :
MSMEG_1359|M.smegmatis_MC2_155 VTTGRRWYLMAYDRDREDWRSLRLDRMS-DVRATG--TTFTARPAPDAAA
TH_1858|M.thermoresistible__bu VTTGRRWYLLAYDRDRGDWRSLRLDRMT-DVRALG--STFTPRPAPDAAE
Mflv_5106|M.gilvum_PYR-GCK VTTGRRWYLMCFDRDRDDWRSLRLDRMS-DVRAVG--TTFIPREAPDAAS
MAB_2185|M.abscessus_ATCC_1997 ITDRGRWYLVGHDRDRDATRTFRLSRIGEDVTKIEKPGTIRRPEGADLRA
MAV_2407|M.avium_104 VTQKGRWYLVGHDRDRDATRTFRLSRIGPGVTPIGPPGAVTVPDGVDLRR
Mvan_2933|M.vanbaalenii_PYR-1 VAKRPTWYLVSN--TDAGRRTFRIDRVS-SVELTD--DAVQRPQGFDLAE
:: ***: *::*:.*: .* :. . *
MSMEG_1359|M.smegmatis_MC2_155 YVGRAISAS---AYPYVARVRYFAPEKVVAQRFPPGTATFEPDGPDA-CI
TH_1858|M.thermoresistible__bu FVRHSISAS---PYRHVARVRFRAPERVVSQHFPAAAVMIEPDGPDT-CI
Mflv_5106|M.gilvum_PYR-GCK YVRRAISSS---PYPYVARVRYRAPADVVARVFPAASVDVEPDGPDA-CI
MAB_2185|M.abscessus_ATCC_1997 IVAAAVGDG---PVRITARVWIAEGRAQELRRMGVVVDSRRLGERDG-DI
MAV_2407|M.avium_104 IVAQTVTDVSATPAGGQARVWVADGRATALRRAGRPVGTRQLGGRDG-EV
Mvan_2933|M.vanbaalenii_PYR-1 SWREIADEVDRKRTPLEAQAMCAADGLGLLRMVLGGRLEVGGSTPDGRIE
*:. : . *
MSMEG_1359|M.smegmatis_MC2_155 VTSGAEYPEQLAMYFATVGHDFEVLEPAEVIDAVGAMADRLRRAVR----
TH_1858|M.thermoresistible__bu VTAGADDPQRIVLHLAAPGLPFEVLEPAEVAEAAAAVADLLTRASGRT--
Mflv_5106|M.gilvum_PYR-GCK VTVGADDPQRMVPWLAMPGCEFEVLEPDEVVDAVRVVAERIARAGAGGD-
MAB_2185|M.abscessus_ATCC_1997 VEIDVASPDRLVRSIASHGADAVALEPAQVRDEVVARLQEQAGVTA----
MAV_2407|M.avium_104 IELDIRATDQLARDIAGYGADAVVLEPQSLRDDVLARLRAHAGADAVGAG
Mvan_2933|M.vanbaalenii_PYR-1 IVIRGHNEYALAGELAGLIEWLEVTGPQGVRDHLASIGGALVARYS----
: :. :* . * : :
MSMEG_1359|M.smegmatis_MC2_155 ----------
TH_1858|M.thermoresistible__bu ----------
Mflv_5106|M.gilvum_PYR-GCK ----------
MAB_2185|M.abscessus_ATCC_1997 ----------
MAV_2407|M.avium_104 RPGGRGGVSA
Mvan_2933|M.vanbaalenii_PYR-1 ----------