For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TQRVIDSLAEFAEQLRGQAEEAEKIGKLTDDTVKTMKQIGSIRLLQPKKHGGMEVHPREFAETVMATAAL DPSAGWINGVVGVHPYQLAYADPRVGAEIWADDVDTWVASPYAPQGVATPVDGGYLFNGRWQFSSGTDHC DWIFLGAMIGDAEGKPLMPPQMLHMILPRKDYEIVEDSWDVVGLRGTGSKDVIVRDAFVPAYRTMDAMKV MDGTAQREAGMTETLYLMPWSTMFPLGISSATIGIAEGALAAALDYQRERVNSSGVAIKDDPYVMYAIGE AAADINAARQEILANADRIYDMVDAGKEVSFQDRAAGRRTQVRAVWRAVSAVDEVFARCGGNGTRMDKPL QRYWRDVHVGQAHAIHVPGTVFHASALSSLGVDPQGPLRAMI*
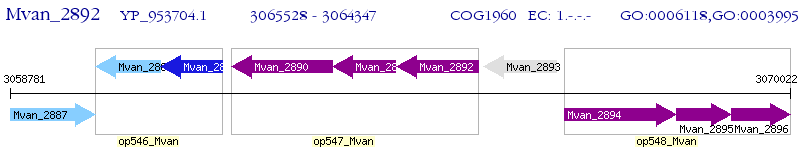
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_2892 | - | - | 100% (393) | acyl-CoA dehydrogenase type 2 |
| M. vanbaalenii PYR-1 | Mvan_5308 | - | 1e-80 | 38.60% (386) | acyl-CoA dehydrogenase type 2 |
| M. vanbaalenii PYR-1 | Mvan_1900 | - | 2e-35 | 29.97% (357) | acyl-CoA dehydrogenase type 2 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3601c | - | 1e-80 | 38.70% (385) | oxidoreductase |
| M. gilvum PYR-GCK | Mflv_4548 | - | 0.0 | 80.41% (393) | acyl-CoA dehydrogenase type 2 |
| M. tuberculosis H37Rv | Rv3570c | - | 2e-80 | 38.70% (385) | oxidoreductase |
| M. leprae Br4923 | MLBr_00737 | fadE25 | 7e-05 | 26.77% (127) | putative acyl-CoA dehydrogenase |
| M. abscessus ATCC 19977 | MAB_0075 | - | 0.0 | 76.08% (393) | putative hydroxylase |
| M. marinum M | MMAR_5065 | - | 4e-77 | 39.02% (369) | acyl-CoA dehydrogenase |
| M. avium 104 | MAV_3020 | - | 0.0 | 83.50% (394) | pigment production hydroxylase |
| M. smegmatis MC2 155 | MSMEG_2892 | - | 0.0 | 91.60% (393) | pigment production hydroxylase |
| M. thermoresistible (build 8) | TH_3059 | - | 0.0 | 88.30% (393) | PUTATIVE pigment production hydroxylase |
| M. ulcerans Agy99 | MUL_4141 | - | 5e-77 | 39.02% (369) | acyl-CoA dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3601c|M.bovis_AF2122/97 -----------MTSIQQRDAQSVLAAIDDLLPEIRDRAQATEDLRRLPDE
Rv3570c|M.tuberculosis_H37Rv -----------MTSIQQRDAQSVLAAIDNLLPEIRDRAQATEDLRRLPDE
MMAR_5065|M.marinum_M -----------MTTIQQRDAQSVLAAIDDLLPQLRERAQATEDLRRLPDE
MUL_4141|M.ulcerans_Agy99 -----------MTTIQQRDAQSVLAAIDDLLPQLRERAQATEDLRRLPDE
MSMEG_2892|M.smegmatis_MC2_155 --------------MTKR----VIDGIAELADQLREQAVEAERIGKLTDT
TH_3059|M.thermoresistible__bu --------------MTKR----VIDGIAELADQLREQSIEAERIGKLTDQ
Mvan_2892|M.vanbaalenii_PYR-1 --------------MTQR----VIDSLAEFAEQLRGQAEEAEKIGKLTDD
MAV_3020|M.avium_104 --------------MSER----VIDRVMAMADQLRDQAAEAEQLGRLTDD
Mflv_4548|M.gilvum_PYR-GCK --------------MSER----VLDRLNELAGQFKEQAVEAEKLGKLPDA
MAB_0075|M.abscessus_ATCC_1997 --MEGQRGGRTIVAMTER----VIDRINDLAEQLREQAWEAEKLGRLPDE
MLBr_00737|M.leprae_Br4923 MVGWSGNPLFDLFKLPEEHNELRATIRALAEKEIAPHAADVDQRARFPEE
: :. :: :: .: ::.:
Mb3601c|M.bovis_AF2122/97 TVKALDDVGFFTLLQPQQWGGLQCDPALFFEATRRLASVCGSTGWVSSIV
Rv3570c|M.tuberculosis_H37Rv TVKALDDVGFFTLLQPQQWGGLQCDPALFFEATRRLASVCGSTGWVSSIV
MMAR_5065|M.marinum_M TVQALEDVGFFTLLQPEQWGGLQCDPTLFYEATRRLASACGSTGWVSSII
MUL_4141|M.ulcerans_Agy99 TVQALEDVGFFTLLQPEQWGGLQCDPTLFYEATRRLASACGSTGWVSSII
MSMEG_2892|M.smegmatis_MC2_155 TVKTMKQVGSIRLLQPKKHGGLEVHPREFAETVMATAALDPSAGWINGVV
TH_3059|M.thermoresistible__bu TVKIMKKLGSIRLLQPKKHGGLEVHPREFAETVMATAALDPSAGWINGVV
Mvan_2892|M.vanbaalenii_PYR-1 TVKTMKQIGSIRLLQPKKHGGMEVHPREFAETVMATAALDPSAGWINGVV
MAV_3020|M.avium_104 TVKLMKGAGLIRLLQTKQYQGFEVHPREFAETVMATAALDPAAGWIVGVV
Mflv_4548|M.gilvum_PYR-GCK TVKSMKSIGSIRLLQPKKHGGLEVHPREFAETVMATAALDPAAGWVNGVV
MAB_0075|M.abscessus_ATCC_1997 TAKMLKAAGLIRLLQPQKYGGYEAHPREFAETVMTTAALDPASGWICGVV
MLBr_00737|M.leprae_Br4923 ALAALNASGFNAIHVPEEYGGQGADSVAACIVIEEVARVDASASLIPAVN
: :. * : .:: * .. . * ::. : .:
Mb3601c|M.bovis_AF2122/97 GVHNWHLALFDQR-AQEEVWGEDPSTRISSSYAPMGAGVVVDGG------
Rv3570c|M.tuberculosis_H37Rv GVHNWHLALFDQR-AQEEVWGEDPSTRISSSYAPMGAGVVVDGG------
MMAR_5065|M.marinum_M GVHNWHLALFDQL-AQEEVWGSDPKTRISSSYAPMGAGVVVDGG------
MUL_4141|M.ulcerans_Agy99 GVHNWHLALFDQL-AQEEVWGSDPKTRISSSYAPMGAGVVVDGG------
MSMEG_2892|M.smegmatis_MC2_155 GVHPYQLAYADPR-VGEEIWAQDVDTWVASPYAPQGVAVPVDGG------
TH_3059|M.thermoresistible__bu GVHPYQLAYADPR-VAEEIWAEDVDTWIASPYAPQGVARPVDGG------
Mvan_2892|M.vanbaalenii_PYR-1 GVHPYQLAYADPR-VGAEIWADDVDTWVASPYAPQGVATPVDGG------
MAV_3020|M.avium_104 GVHPYQLAYADPK-VAAEVWADDVDTWMASPYAPQGVAKPVNGG------
Mflv_4548|M.gilvum_PYR-GCK GVHPYQLAYADPK-VGEEIWADDIDTWVASPYAPQGVAVPTDGG------
MAB_0075|M.abscessus_ATCC_1997 GVHPWQVAFADSR-VAEEMWGEDQDIWMASPYAPTGVAKPVDGG------
MLBr_00737|M.leprae_Br4923 KLGTMGLILRGSEELKKQVLPSLAAEGAMASYALSEREAGSDAASMRTRA
: : . :: . :.** :..
Mb3601c|M.bovis_AF2122/97 ------YLVNGSWNWSSGCDHASWTFVGGPVIKDGR-----PVDFGSFLI
Rv3570c|M.tuberculosis_H37Rv ------YLVNGSWNWSSGCDHASWTFVGGPVIKDGR-----PVDFGSFLI
MMAR_5065|M.marinum_M ------YLVNGSWNWSSGCDHATWCFAGGPVIKDGR-----PVDFGSFLI
MUL_4141|M.ulcerans_Agy99 ------YLVNGSWNWSSGCDHGTWCFAGGPVIKDGR-----PVDFGSFLI
MSMEG_2892|M.smegmatis_MC2_155 ------YIFNGRWQFSSGTDHCDWIILG-AMVGDADGTPLMPPQMLHMIL
TH_3059|M.thermoresistible__bu ------YLFNGRWQFSSGTDHCDWIILG-AMVGDADGTPVTPPRMLHMIL
Mvan_2892|M.vanbaalenii_PYR-1 ------YLFNGRWQFSSGTDHCDWIFLG-AMIGDAEGKPLMPPQMLHMIL
MAV_3020|M.avium_104 ------YIFNGRWQFSSGTDHCDWIILG-AMLGDEQGVPLMPPQMLHMIL
Mflv_4548|M.gilvum_PYR-GCK ------YIFTGRWQFSSGTDACDWIILG-AMVGDAKGVPLMPPQMLHMIL
MAB_0075|M.abscessus_ATCC_1997 ------YIFNGRWQFSSGTDHCDWIILG-AMLGDADGTPIMPPKMLHMIL
MLBr_00737|M.leprae_Br4923 KADGDDWILNGFKCWITNGGKSTWYTVMAVTDPDKG-----ANGISAFIV
::..* : :. . * * . : :::
Mb3601c|M.bovis_AF2122/97 PRSEYEI-KDVWY-VVGLRGTGSNTLVVKDVFVPRHRFLSYKAMNDHTAG
Rv3570c|M.tuberculosis_H37Rv PRSEYEI-KDVWY-VVGLRGTGSNTLVVKDVFVPRHRFLSYKAMNDHTAG
MMAR_5065|M.marinum_M PRTEYRI-DDVWH-VVGLRGTGSNTVVVKDVFVPRHRFLSYKAMNDHTAG
MUL_4141|M.ulcerans_Agy99 PRTEYRI-DDVWH-VVGLRGTGSNTVVVKDVFVPRHRFLSYKAMNDHTAG
MSMEG_2892|M.smegmatis_MC2_155 PRKDYEIVDDSWD-VVGLRGTGSKDVIVKDAFVPAYRTMDGMKVMDGTAQ
TH_3059|M.thermoresistible__bu PRKDYEIVEDSWD-VVGLRGTGSKDVIVKDAFVPAYRTMDGMKVMDGTAQ
Mvan_2892|M.vanbaalenii_PYR-1 PRKDYEIVEDSWD-VVGLRGTGSKDVIVRDAFVPAYRTMDAMKVMDGTAQ
MAV_3020|M.avium_104 PRKDYEIVEDSWN-VVGLRGTGSKDVIVKDAFVPSYRTMDATKVMDGTAQ
Mflv_4548|M.gilvum_PYR-GCK PRKDYQIVDDSWD-VVGLRGTGSKDVIVDGAFVPTYRTMDGMKVMDGSAQ
MAB_0075|M.abscessus_ATCC_1997 PRADYQIVEDSWN-VVGLRGTGSKDIIVNDAFVPSYRVMDGDQVIDGTAQ
MLBr_00737|M.leprae_Br4923 HKDDEGFSIGPKEKKLGIKGSPTTELYFDKCRIPGDRIIGEPGTGFKTAL
: : : . :*::*: :. : . :* * :. :*
Mb3601c|M.bovis_AF2122/97 GLATNSAPVYKMPWGTMHPTTISAPIVGMAYGAYAAHVEHQGKRVRAAFA
Rv3570c|M.tuberculosis_H37Rv GLATNSAPVYKMPWGTMHPTTISAPIVGMAYGAYAAHVEHQGKRVRAAFA
MMAR_5065|M.marinum_M GLQTNTAPVYKMPWGTMHPTTITAPIVGMAYGAYDAHVEHQGKRVRAAFA
MUL_4141|M.ulcerans_Agy99 GLQTNTAPVYKMPWGTMHPTTITAPIVGMAYGAYDAHVEHQGKRVRAAFA
MSMEG_2892|M.smegmatis_MC2_155 REAGMTEPLYLMPWSTMFPLGISSATIGIAEGALAAALDYQRQRVNS--S
TH_3059|M.thermoresistible__bu REAGMTEPLYLMPWSTMFPLGISSATIGIAEGALAAALDYQRRRVNS--S
Mvan_2892|M.vanbaalenii_PYR-1 REAGMTETLYLMPWSTMFPLGISSATIGIAEGALAAALDYQRERVNS--S
MAV_3020|M.avium_104 REAGMTEPLYLMPWSTMFPLGISAATIGIAEGALAAHLDYQRSRVGA--T
Mflv_4548|M.gilvum_PYR-GCK IEAGMTEPLYKMPWSTMFPLGISSATIGICEGALAAHLDYQRERVGA--N
MAB_0075|M.abscessus_ATCC_1997 RDAGMTDTLYRMPWSTMFPLGISSAVIGIAEGALAAHLEYQRERVGA--Q
MLBr_00737|M.leprae_Br4923 ATLDHTRP------------TIGAQAVGIAQGALDAAIVYTKDRKQF---
: . * : :*:. ** * : : *
Mb3601c|M.bovis_AF2122/97 GEKAKDDPFAKVRIAEAASDIDAAWRQLIGNVSDEYALLAAGKEIPFELR
Rv3570c|M.tuberculosis_H37Rv GEKAKDDPFAKVRIAEAASDIDAAWRQLIGNVSDEYALLAAGKEIPFELR
MMAR_5065|M.marinum_M GEKAKDDPFAKVRIAEAASDIDAAWRQLMGNVGDEYALLSAGKEIPFDLR
MUL_4141|M.ulcerans_Agy99 GEKAKDDPFAKVRIAEAASDIDAAWRQLMGNVGDEYALLSAGKEIPFDLR
MSMEG_2892|M.smegmatis_MC2_155 GVAVKDDPYVMYAIGEAAADINAARQELLANADRIYDMVAAGKEVSFEDR
TH_3059|M.thermoresistible__bu GVAVKDDPYVMYAIGEAAADINAARQELLANADRIYDIVASGAEVSFEDR
Mvan_2892|M.vanbaalenii_PYR-1 GVAIKDDPYVMYAIGEAAADINAARQEILANADRIYDMVDAGKEVSFQDR
MAV_3020|M.avium_104 GTAIKDDPYVMYAIGEAAADINAARQELLANADRIYDIVASGKEVSFVDR
Mflv_4548|M.gilvum_PYR-GCK GTAIKDDPYVMYAIGDAAAEIEASRNTLLSNVDRVYDIVDSGREVTFETR
MAB_0075|M.abscessus_ATCC_1997 GTAVKDDPYVLFAIGEAAADINAARQEILANVDKIWGIVDSGKEVSFADR
MLBr_00737|M.leprae_Br4923 GESISTFQSIQFMLADMAMKVEAARLIVYAAAARAE---RGEPDLGFISA
* . :.: * .::*: : . . . :: *
Mb3601c|M.bovis_AF2122/97 ARARRDQVRATGRSIASIDRLFEASGATALSNEAPIQRFWRDAHAGRVHA
Rv3570c|M.tuberculosis_H37Rv ARARRDQVRATGRSIASIDRLFEASGATALSNEAPIQRFWRDAHAGRVHA
MMAR_5065|M.marinum_M TRARRDQVRATGRSIASIDRLFEASGATALANDAPIQRFWRDAHAGRVHA
MUL_4141|M.ulcerans_Agy99 TRARRDQVRATGRSIASIDRLFEASGATALANDAPIQRFWRDAHAGRVHA
MSMEG_2892|M.smegmatis_MC2_155 AAGRRTQIRAVWRAVSAVDEIFARCGGNAARMDKPLQRYWRDVHVGQAHA
TH_3059|M.thermoresistible__bu AAGRRTQVRAVWRAVSAVDEIFARCGGNAARMDQPLQRFWRDVHVAQAHA
Mvan_2892|M.vanbaalenii_PYR-1 AAGRRTQVRAVWRAVSAVDEVFARCGGNGTRMDKPLQRYWRDVHVGQAHA
MAV_3020|M.avium_104 AAGRRTQVRAVWRAVSAVDEIFARSGGNAARMDKPLQRYWRDVHVGQMHA
Mflv_4548|M.gilvum_PYR-GCK AAVRRSQIKAVWRAVAAVDQIFARSGGNALRMDKPLQRYWRDAHAGVAHA
MAB_0075|M.abscessus_ATCC_1997 AAGRRTQVRAAWRAVMAVDQIFSRSGGNAMRLDKPLQRYWRDAHTGLAHA
MLBr_00737|M.leprae_Br4923 AS----KCFASDIAMEVTTDAVQLFGGAGYTSDFPVERFMRDAKITQIYE
: : * :: . *. . : *::*: **.: :
Mb3601c|M.bovis_AF2122/97 ANDPERAYVIFGNHEFGLPP----GDTMV-
Rv3570c|M.tuberculosis_H37Rv ANDPERAYVIFGNHEFGLPP----GDTMV-
MMAR_5065|M.marinum_M ANDPERAYVIFGNNEFGLPA----GDTMV-
MUL_4141|M.ulcerans_Agy99 ANDPERAYVIFGNNEFGLPA----GDTMV-
MSMEG_2892|M.smegmatis_MC2_155 IHVPGTVYHASALSSLGVDP-QGPLRAMI-
TH_3059|M.thermoresistible__bu IHSPGTVYHASALSSLGVEP-QGPLRAMI-
Mvan_2892|M.vanbaalenii_PYR-1 IHVPGTVFHASALSSLGVDP-QGPLRAMI-
MAV_3020|M.avium_104 IHVPGTTYHASALSSIGIDPPEGPLRALI-
Mflv_4548|M.gilvum_PYR-GCK IHVPGTVYHASALSSLGVDP-QGPLRAMI-
MAB_0075|M.abscessus_ATCC_1997 IHVPSTVFHASALSSLGIEP-QGPLRSMI-
MLBr_00737|M.leprae_Br4923 GTNQIQRVVMS--------------RALLR
:::