For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TDRIEVPRTIAAPASEIFSVLRDPQGHVAIDATGMLQDADGDPVRAVGDTFVVHMDREALNDFPELGRYD VTVQITEYEQDSLIAWTILGRLRPQIGHVYGYRLEPAEQGTLVTSFYDWSGIHQQWREAGIFPVVSELAL RGTLGILDRTVRRGYRRG*
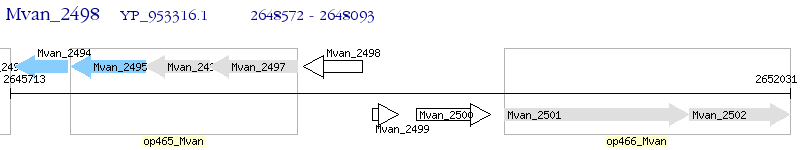
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_2498 | - | - | 100% (159) | hypothetical protein Mvan_2498 |
| M. vanbaalenii PYR-1 | Mvan_0758 | - | 2e-13 | 34.97% (143) | hypothetical protein Mvan_0758 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0091 | - | 4e-05 | 25.89% (112) | hypothetical protein Mb0091 |
| M. gilvum PYR-GCK | Mflv_3906 | - | 2e-76 | 84.18% (158) | hypothetical protein Mflv_3906 |
| M. tuberculosis H37Rv | Rv0088 | - | 4e-05 | 25.89% (112) | hypothetical protein Rv0088 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_0253 | - | 2e-06 | 31.40% (121) | hypothetical protein MMAR_0253 |
| M. avium 104 | MAV_5230 | - | 8e-08 | 28.93% (121) | hypothetical protein MAV_5230 |
| M. smegmatis MC2 155 | MSMEG_4744 | - | 2e-12 | 32.08% (159) | hypothetical protein MSMEG_4744 |
| M. thermoresistible (build 8) | TH_4053 | - | 1e-08 | 31.54% (149) | PUTATIVE conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_4861 | - | 1e-06 | 31.40% (121) | hypothetical protein MUL_4861 |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_2498|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_3906|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_4744|M.smegmatis_MC2_155 --------------------------------------------------
TH_4053|M.thermoresistible__bu --------------------------------------------------
MMAR_0253|M.marinum_M --------------------------------------------------
MUL_4861|M.ulcerans_Agy99 --------------------------------------------------
MAV_5230|M.avium_104 --------------------------------------------------
Mb0091|M.bovis_AF2122/97 MSVYKHAPSRVRLRQTRSTVVKGRSGSLSWRRVRTGDLGLAVWGGREEYR
Rv0088|M.tuberculosis_H37Rv MSVYKHAPSRVRLRQTRSTVVKGRSGSLSWRRVRTGDLGLAVWGGREEYR
Mvan_2498|M.vanbaalenii_PYR-1 -----------------------MTDRIEVPRTIAAPASEIFSVLRDPQG
Mflv_3906|M.gilvum_PYR-GCK -----------------------MTDRIEVSRIIAAPPADIFGVLCDPQG
MSMEG_4744|M.smegmatis_MC2_155 ----------------MS-----TTKRYVVTRRIHAAPQDVFRVLADPAR
TH_4053|M.thermoresistible__bu ------------MSDIQT-----TGTSLAVHTTVNAPARTVFDVLADPTT
MMAR_0253|M.marinum_M ------------MVDTDAPLPDRSERVVSATREIAAGAAQIFELIADPSR
MUL_4861|M.ulcerans_Agy99 ------------MVDTDAPLPDRSERVVSATREIAAGAAQIFELIADPSR
MAV_5230|M.avium_104 ------------MSDTEA------ERIVSATRVIAANAARIFELIADPAR
Mb0091|M.bovis_AF2122/97 AVKPGTPGIQPKGDMMTVTVVDAGPGRVSRSVEVAAPAAELFAIVADPRR
Rv0088|M.tuberculosis_H37Rv AVKPGTPGIQPKGDMMTVTVVDAGPGRVSRSVEVAAPAAELFAIVADPRR
: * . :* :: **
Mvan_2498|M.vanbaalenii_PYR-1 HVAIDATGMLQD-ADGDPVRAVGDTFVVHMDREALNDFPELGRYDVTVQI
Mflv_3906|M.gilvum_PYR-GCK HVAIDATGMLQD-ADGSPARAIGDSFVVHMDRAALNDFPELGRYDVTVAI
MSMEG_4744|M.smegmatis_MC2_155 HKDTEPGDWVRDAIDPQPITDTGQIFAINMFLEQAGG-----HYVMHNRV
TH_4053|M.thermoresistible__bu HARIDGTGWVREPVDGARLTGAGQIFRMGMYHENHPHR----HYEIANRV
MMAR_0253|M.marinum_M QPDWDGNDNLAHCAAGQRVHRVGDVFRMTLTLGSVREN----------HI
MUL_4861|M.ulcerans_Agy99 QPDWDGNDNLAHCAAGQRVHRVGDVFRMTLTLGSVREN----------HI
MAV_5230|M.avium_104 QPGWDGNDNLASAAPGQRVHRVGDVFKMTLTNGAVREN----------HV
Mb0091|M.bovis_AF2122/97 HRELDGSGTVRGNIKVPAKLVVGSKFSTKMKLFGLPYR-------ITSRV
Rv0088|M.tuberculosis_H37Rv HRELDGSGTVRGNIKVPAKLVVGSKFSTKMKLFGLPYR-------ITSRV
: : . : *. * : :
Mvan_2498|M.vanbaalenii_PYR-1 TEYEQDSLIAWTILG-------------RLRPQIGHVYGYRLEPAEQ---
Mflv_3906|M.gilvum_PYR-GCK TQYEQDALIAWTILG-------------QLKPQIGHVYGYRLEPADT---
MSMEG_4744|M.smegmatis_MC2_155 TEYDEDRTIAWEPG---------LLDDAGNHSPGGWFWRYDLQPDGD---
TH_4053|M.thermoresistible__bu EVFDPPHAIAWQPGAEPRHIPGRGADATGPVEFGGWIWRYDLAPQHAPGA
MMAR_0253|M.marinum_M VEFTEARLIAWRPSE-------------LGKTPPGHLWRWELTPISG---
MUL_4861|M.ulcerans_Agy99 VEFTEARLIAWRPSE-------------LGKTPPGHLWRWELTPISG---
MAV_5230|M.avium_104 TEFVEGSVIAWRPAE-------------PGKQPPGHLWRWELSPVNA---
Mb0091|M.bovis_AF2122/97 TALKPNELVEWSHPL-------------------GHRWRWEFESLSP---
Rv0088|M.tuberculosis_H37Rv TALKPNELVEWSHPL-------------------GHRWRWEFESLSP---
: * * : : : .
Mvan_2498|M.vanbaalenii_PYR-1 --GTLVTSFYDWSGIHQQWR-----EAGIFPVVSELALRGTLGILDRTVR
Mflv_3906|M.gilvum_PYR-GCK --GTLVTSFYDWSDIHQHWR-----DMGIFPVISELALRGTLGILDRTVR
MSMEG_4744|M.smegmatis_MC2_155 --ATAVTLTYDWTDTPQSFAD----QIGGMPPFPEQFIAESLATLDRAVR
TH_4053|M.thermoresistible__bu PAGTRVTLTYDWSQVPAADRR----AMR-FPPFPPEHLVNSLRHLKELAE
MMAR_0253|M.marinum_M -TRTLVTHTYDWTELSDPAR------FPRARATTTVQLRASLDRLAGHAE
MUL_4861|M.ulcerans_Agy99 -TRTLVTHTYDWTELSDPAR------FPRARATTTVQLRASLDRLAGHAE
MAV_5230|M.avium_104 -ELTTVTHTYDWTALTDPTR------LQRARATTPEMLRESMDRLA---T
Mb0091|M.bovis_AF2122/97 -TLTRVTETFDYHAAGAIKNGLKFYEMTGFAKSNAAGIEATLAKLSDQYA
Rv0088|M.tuberculosis_H37Rv -TLTRVTETFDYHAAGAIKNGLKFYEMTGFAKSNAAGIEATLAKLSDQYA
* ** :*: : :: *
Mvan_2498|M.vanbaalenii_PYR-1 RGYRRG
Mflv_3906|M.gilvum_PYR-GCK RGYPRS
MSMEG_4744|M.smegmatis_MC2_155 ------
TH_4053|M.thermoresistible__bu RDVPR-
MMAR_0253|M.marinum_M AAAAGD
MUL_4861|M.ulcerans_Agy99 AAAAGD
MAV_5230|M.avium_104 LAQSGD
Mb0091|M.bovis_AF2122/97 RGRA--
Rv0088|M.tuberculosis_H37Rv RGRA--