For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SPKSLSARVVGALALAAALPFAATACGGGDGGGGGAGGGGDTIVIGTKFDQPGLGQKNPDGTMSGFDVDV ATYVAGELGYTPDKIEWKESPSAQRENLIQNGQVTFIAATYSITDARKEKVSFAGPYLITGQSLLVRSDN TDITGEASLENNKILCSVSGSTPAQKIKDKYQGVQLQQYDTYSACIDALKNGAVDAVTTDEVILAGYAAQ SPGAFKIVGEPFSEERYGIGLKKDDTELRTKINDALKKMEESGAWKEAFDKNLGPAGITAPTPPPLDN*
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
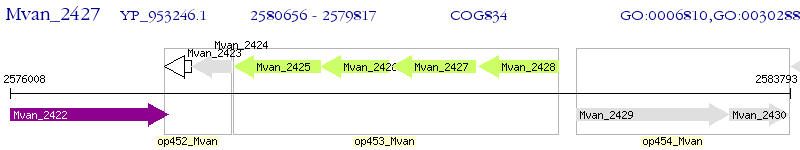
| M. vanbaalenii PYR-1 | Mvan_2427 | - | - | 100% (279) | extracellular solute-binding protein |
| M. vanbaalenii PYR-1 | Mvan_0704 | - | 1e-27 | 34.11% (214) | extracellular solute-binding protein |
| M. vanbaalenii PYR-1 | Mvan_4703 | - | 2e-18 | 28.03% (264) | polar amino acid ABC transporter, inner membrane subunit |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0419c | glnH | 3e-26 | 31.78% (214) | glutamine-binding lipoprotein |
| M. gilvum PYR-GCK | Mflv_3969 | - | 1e-148 | 91.76% (279) | extracellular solute-binding protein |
| M. tuberculosis H37Rv | Rv0411c | glnH | 3e-26 | 31.78% (214) | glutamine-binding lipoprotein |
| M. leprae Br4923 | MLBr_00303 | glnH | 4e-25 | 28.76% (233) | putative glutamine-binding protein |
| M. abscessus ATCC 19977 | MAB_3051 | - | 2e-63 | 47.37% (285) | putative glutamate ABC transporter, periplasmic protein |
| M. marinum M | MMAR_0714 | glnH | 3e-25 | 31.31% (214) | glutamine-binding lipoprotein GlnH |
| M. avium 104 | MAV_4750 | - | 7e-24 | 30.37% (214) | extracellular solute-binding protein, family protein 3 |
| M. smegmatis MC2 155 | MSMEG_2727 | - | 1e-128 | 81.09% (275) | glutamate binding protein |
| M. thermoresistible (build 8) | TH_1785 | - | 1e-106 | 70.30% (266) | glutamate binding protein |
| M. ulcerans Agy99 | MUL_2809 | glnH | 2e-25 | 31.31% (214) | glutamine-binding lipoprotein GlnH |
CLUSTAL 2.0.9 multiple sequence alignment
MLBr_00303|M.leprae_Br4923 --------MMTRQFKIQRACS---VLATAMMLAACGHAELLTVASMPTLP
MAV_4750|M.avium_104 MAEAERVPMTAGLPLLRRACT---ALAAVLVAAGCGHTESLRVASVPTLP
Mb0419c|M.bovis_AF2122/97 ---------MTRRALLARAAAPLAPLALAMVLASCGHSETLGVEATPTLP
Rv0411c|M.tuberculosis_H37Rv ---------MTRRALLARAAAPLAPLALAMVLASCGHSETLGVEATPTLP
MMAR_0714|M.marinum_M ---------MRRLPGLRRAASALAVLALVITLDGCGHSEPLKVESTPTLP
MUL_2809|M.ulcerans_Agy99 ---------MRRLPGLRRAASALAVLALVITLDGCGHSEPLKVESTPTLP
Mvan_2427|M.vanbaalenii_PYR-1 ------------------------------MSPKSLSARVVG---ALALA
Mflv_3969|M.gilvum_PYR-GCK ------------------------------MFPKSTSTRIAG---ALALA
MSMEG_2727|M.smegmatis_MC2_155 --------------------------------MRSIPKRVAG---AVALA
TH_1785|M.thermoresistible__bu ----------------------------MPFTSTSLLGRCLRRGAAVALA
MAB_3051|M.abscessus_ATCC_1997 -----------------------------MSRLRDEDARVMRR-RHTALV
. :*
MLBr_00303|M.leprae_Br4923 PPTPVGMEQVPPELLLPQDNTEEDCDPTASLRPFPTKAEADAAVAEIRAR
MAV_4750|M.avium_104 PPTPVGMEQLPPQPPLPPDGPDQNCDLTASLRPFPTKAEADAAVADIRAR
Mb0419c|M.bovis_AF2122/97 LPTPVGMEIMPPQPPLPPDSSSQDCDPTASLRPFATKAEADAAVADIRAR
Rv0411c|M.tuberculosis_H37Rv LPTPVGMEIMPPQPPLPPDSSSQDCDPTASLRPFATKAEADAAVADIRAR
MMAR_0714|M.marinum_M LPTPVGMEVMPPEPPLPADSSSQDCDPTASLRPFPTKAEADAAVADIRAR
MUL_2809|M.ulcerans_Agy99 LPTPVGMEVMPPEPPLPADSSSQDCDPTASLRPFPTKAEADAAVADIRAR
Mvan_2427|M.vanbaalenii_PYR-1 AALPFAAT--------------------------ACGGGDGGGGGAGGGG
Mflv_3969|M.gilvum_PYR-GCK AALPFAMT--------------------------ACGGGGDSGGGGGAGG
MSMEG_2727|M.smegmatis_MC2_155 IALPFAAT--------------------------ACGGGD-----SGGDS
TH_1785|M.thermoresistible__bu VGLALTAT--------------------------ACGADR--------DE
MAB_3051|M.abscessus_ATCC_1997 AALALLATLVS-----------------------ACGSTEPRQLLGSIRG
.. . .
MLBr_00303|M.leprae_Br4923 GRLIVGLEIGSNLFSFRDPITGEITGFDIDIAGEVAR----DIFGTPSHM
MAV_4750|M.avium_104 GRLIVGLDIGSNLFSFRDPITGEITGFDVDIAGEIAR----DIFGAPSHV
Mb0419c|M.bovis_AF2122/97 GRLIVGLDIGSNLFSFRDPITGEITGFDVDIAGEVAR----DIFGVPSHV
Rv0411c|M.tuberculosis_H37Rv GRLIVGLDIGSNLFSFRDPITGEITGFDVDIAGEVAR----DIFGVPSHV
MMAR_0714|M.marinum_M GRLIVGLDIGSNLFSFRDPITGEITGFDVDIASEVAR----DIFGGPTHV
MUL_2809|M.ulcerans_Agy99 GRLIVGLDIGSNLFSFRDPITGEITGFDVDIASEVAR----DIFGGPTHV
Mvan_2427|M.vanbaalenii_PYR-1 DTIVIGTKFDQPGLGQKNPD-GTMSGFDVDVATYVAG----ELGYTPDKI
Mflv_3969|M.gilvum_PYR-GCK DTIVIGTKFDQPGLGQKNPD-GTMSGFDVDVATYVAG----ELGYAPDKI
MSMEG_2727|M.smegmatis_MC2_155 DTIVIGTKYDQPGLGLKQPD-GTLTGFDVDVAKYVAN----ELGYSEDQI
TH_1785|M.thermoresistible__bu NTVVIGTKFDQPGLAMKNED-GSMSGFDVAVAEYVAQ----ELGYAPEDI
MAB_3051|M.abscessus_ATCC_1997 GTVILGTKFDQPGVGLQHPD-KKMSGFDVKISEYVVNAIADELGVKHPTI
. :::* . .. .. :. ::***: :: :. :: :
MLBr_00303|M.leprae_Br4923 EYRILSTAARITALQKSEVDIIVKTMTITCKRRKLVNFSTVYLDAKQRIL
MAV_4750|M.avium_104 EYRILSSDERVTALQRGEVDVVVKTMTITCDRRKQVNFSTVYLDANQRIL
Mb0419c|M.bovis_AF2122/97 EYRILSAAERVTALQKSQVDIVVKTMSITCERRKLVNFSTVYLDANQRIL
Rv0411c|M.tuberculosis_H37Rv EYRILSAAERVTALQKSQVDIVVKTMSITCERRKLVNFSTVYLDANQRIL
MMAR_0714|M.marinum_M EYRILSAAERITALQSSQVDVVVKTMTITCERRKLVNFSTVYLDANQRIL
MUL_2809|M.ulcerans_Agy99 EYRILSAAERITALQSSQVDVVVKTMTITCERRKLVNFSTVYLDANQRIL
Mvan_2427|M.vanbaalenii_PYR-1 EWKESPSAQRENLIQNGQVTFIAATYSITDARKEKVSFAGPYLITGQSLL
Mflv_3969|M.gilvum_PYR-GCK EWKESPSAQRETLIQNGQVTFIAATYSITDSRKEKVAFAGPYLITGQSLL
MSMEG_2727|M.smegmatis_MC2_155 EWKEAPSAQRETLIQNGQVDFIAATYSITDSRKEKVDFAGPYLITGQSLL
TH_1785|M.thermoresistible__bu EWKEAPSNQRETLIQNGQVDFIVGTYSITDSRKERIDFAGPYLITGQSLL
MAB_3051|M.abscessus_ATCC_1997 RWYETPSAQREQLIDNGTVDMIAGSYSVNYSRAQKVSFAGPYLITYQGLL
.: .: * :: . * .:. : ::. * : : *: ** : * :*
MLBr_00303|M.leprae_Br4923 APR-DSPITKPSDLS-NKRVCMTRGTTSLHQVSEVVPPPIIVSAENWADC
MAV_4750|M.avium_104 APR-DSPITKVSDLS-GKRVCVAKGTTSLHRIRQIDPPPIVVSVVNWADC
Mb0419c|M.bovis_AF2122/97 APR-DSPITKVSDLS-GKRVCVARGTTSLRRIREIAPPPVIVSVVNWADC
Rv0411c|M.tuberculosis_H37Rv APR-DSPITKVSDLS-GKRVCVARGTTSLRRIREIAPPPVIVSVVNWADC
MMAR_0714|M.marinum_M TSR-DSPIFKVSDLS-GKRVCVARGTTSLRRIREIAPPPVIVSVVNWADC
MUL_2809|M.ulcerans_Agy99 TSR-DSPIFKVSDLS-GKRVCVARGTTSLRRIREIAPPPVIVSVVNWADC
Mvan_2427|M.vanbaalenii_PYR-1 VRSDNTDITGEASLENNKILCSVSGSTPAQKIKDKYQGVQLQQYDTYSAC
Mflv_3969|M.gilvum_PYR-GCK VRNDNNDITGAASLENNKILCSVSGSTPAQKIKDEYQGVQLQQYDTYSAC
MSMEG_2727|M.smegmatis_MC2_155 VRADNNDITGAASLENNKKLCSVSGSTPAQRIKDEYPGVQLQQYDTYSAC
TH_1785|M.thermoresistible__bu VRADDTDITGPASLENNKLLCSVTGSTPAQRIKDEYPGVQLQEYDTYSAC
MAB_3051|M.abscessus_ATCC_1997 VRKADDSLTTLDDLNRGKKLCSVSGSTPAQNVKNLLPGTQLQEFDSYSSC
. : : .*. .* :* . *:*. :.: : : . .:: *
MLBr_00303|M.leprae_Br4923 LVAMQQREIDAISTDDAILAGLVEEDP-YLHIV-----------------
MAV_4750|M.avium_104 LVAMQQREIDAVSTDDSILAGLVEEDP-YLHIV-----------------
Mb0419c|M.bovis_AF2122/97 LVALQQREIDAVSTDDTILAGLVEEDP-YLHIV-----------------
Rv0411c|M.tuberculosis_H37Rv LVALQQREIDAVSTDDTILAGLVEEDP-YLHIV-----------------
MMAR_0714|M.marinum_M LVALQQREIDAVSTDDTILAGLVEEDP-YLHIV-----------------
MUL_2809|M.ulcerans_Agy99 LVALQQREIDAVSTDDTILAGLVEEDP-YLHIV-----------------
Mvan_2427|M.vanbaalenii_PYR-1 IDALKNGAVDAVTTDEVILAGYAAQSPGAFKIV-----------------
Mflv_3969|M.gilvum_PYR-GCK IDALKNGAVDAVTTDEVILAGYAAQSPGTFKIV-----------------
MSMEG_2727|M.smegmatis_MC2_155 VEALKNGAVDALTTDEVILAGYAAQSPGTFKLV-----------------
TH_1785|M.thermoresistible__bu VEALRNNAVDAVTTDEVILAGYAAQSPDDLRVV-----------------
MAB_3051|M.abscessus_ATCC_1997 VEALRRGKVDALTTDETILAGYAKRYEGEFKLIQMNYELSKPLCIGNPKR
: *::. :**::**: **** . . ::::
MLBr_00303|M.leprae_Br4923 ----GPNMATQPYGIGINLNNTTLVRFVNGTLERIRRDGTWITLYRKWLT
MAV_4750|M.avium_104 ----GPNMATQPYGIGINLNNTGLVRFVNGTLERIRRDGTWNTLYRKWLT
Mb0419c|M.bovis_AF2122/97 ----GPDMADQPYGVGINLDNTGLVRFVNGTLERIRNDGTWNTLYRKWLT
Rv0411c|M.tuberculosis_H37Rv ----GPDMADQPYGVGINLDNTGLVRFVNGTLERIRNDGTWNTLYRKWLT
MMAR_0714|M.marinum_M ----GPNMANQPYGIGINLDNTGLVRFVNGTLERIRNDGTWNTLYRKWLS
MUL_2809|M.ulcerans_Agy99 ----GPNMANQPYGIGINLDNTGLVRFVNGTLERIRNDGTWNTLYRKWLS
Mvan_2427|M.vanbaalenii_PYR-1 ----GEPFSEERYGIGLKKDDTELRTKINDALKKMEESGAWKEAFDKNLG
Mflv_3969|M.gilvum_PYR-GCK ----GEPFSEERYGIGLKKDDAELRTKINDALKKMEDSGAWKEAFDKNLG
MSMEG_2727|M.smegmatis_MC2_155 ----GDTFSEERYGIGLKKDDTELRNKINDAIEKMESSGAWKEAFDKNLG
TH_1785|M.thermoresistible__bu ----GQPFSEERYGIGLAKGDTELKEKINAAIEKMQQDGEWQRAFEVNLG
MAB_3051|M.abscessus_ATCC_1997 QRRNGDPFSREVYGIGLARGDTAAIEAIDRALDKMIATGAWDSDLREALG
* :: : **:*: .:: :: ::.:: * * *
MLBr_00303|M.leprae_Br4923 VLGPAP-----------APPTP------------RYVD------
MAV_4750|M.avium_104 VLGPAP-----------APPTP------------RYLD------
Mb0419c|M.bovis_AF2122/97 VLGPAP-----------APPTP------------RYVD------
Rv0411c|M.tuberculosis_H37Rv VLGPAP-----------APPTP------------RYVD------
MMAR_0714|M.marinum_M VLGPAP-----------APPRP------------RYVD------
MUL_2809|M.ulcerans_Agy99 VLGPAP-----------APPRP------------RYVD------
Mvan_2427|M.vanbaalenii_PYR-1 PAGIT-------------APTP------------PPLDN-----
Mflv_3969|M.gilvum_PYR-GCK PAGIT-------------APAP------------PPLDN-----
MSMEG_2727|M.smegmatis_MC2_155 PAGIQ-------------APQP------------PAVDRD----
TH_1785|M.thermoresistible__bu DAGID-------------TPTP------------PTVDRN----
MAB_3051|M.abscessus_ATCC_1997 DSTIDGWEKRAESGTYGLRPDPSLAAIEKITGTKPAADPACGAA
* * *