For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MASGSSEHATTPLSKSVLGIAIVAITGMQLMSTLDGTIVIVALPRMQAELNLSDAGKSWVITAYVLAFGG LLLLGGRIGDAVGHKRAFLSGVGLFTIASLVCGLATEEITLVAARAFQGAGAAVAAPTGLALIATTYAVG HARNRALAVSAAMLATGSVLGLVLGGALTVVSWRLAFLINVPIGIAIIAIAVFRLAETHHERLKLDVTGA LLATLACTSAVLFFTQGPSRGWADPFVIGAAVASALLFVAFFVVERSADHPIVPLTVFDNRNRVATFLSL FLAGGVMLSATVMIGLYVQDVMGYSALQAGMAFIPFALALGVGTVVAARVAPYLAPRWIILGAGLLVPCA MLYGSTLDRGIPYVPDLIAPLVVGGFGIGVISVILPLCAIADVGAREIGPVSSITLMVQNLGGPVVLVVI QAVQTSRTLYLGGTTGPVAAMTPAQLDALDAGYTYSLLWVAAVSVLVGAAALFIGFSARQIAVAKYNRDA VETGELWISAEQRGATEG
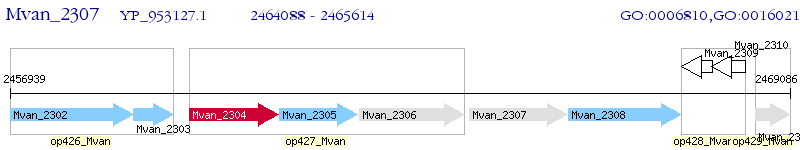
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_2307 | - | - | 100% (508) | major facilitator superfamily transporter |
| M. vanbaalenii PYR-1 | Mvan_2306 | - | e-172 | 63.24% (476) | major facilitator superfamily transporter |
| M. vanbaalenii PYR-1 | Mvan_1284 | - | 4e-43 | 29.95% (434) | major facilitator superfamily transporter |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2871c | efpA | 1e-160 | 58.91% (477) | integral membrane efflux protein EfpA |
| M. gilvum PYR-GCK | Mflv_4046 | - | 1e-169 | 62.18% (476) | EmrB/QacA family drug resistance transporter |
| M. tuberculosis H37Rv | Rv2846c | efpA | 1e-160 | 58.91% (477) | integral membrane efflux protein EfpA |
| M. leprae Br4923 | MLBr_01562 | - | 1e-164 | 59.54% (477) | putative transmembrane efflux protein |
| M. abscessus ATCC 19977 | MAB_3142c | - | 1e-150 | 56.33% (474) | putative transmembrane efflux protein |
| M. marinum M | MMAR_1887 | efpA | 1e-163 | 59.12% (477) | integral membrane efflux protein EfpA |
| M. avium 104 | MAV_3702 | - | 1e-162 | 59.33% (477) | transporter, major facilitator family protein |
| M. smegmatis MC2 155 | MSMEG_2619 | - | 1e-167 | 60.50% (476) | efflux protein |
| M. thermoresistible (build 8) | TH_2858 | - | 1e-172 | 63.45% (476) | PUTATIVE major facilitator superfamily MFS_1 |
| M. ulcerans Agy99 | MUL_2115 | efpA | 1e-162 | 58.70% (477) | integral membrane efflux protein EfpA |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1887|M.marinum_M MTALNDTERAVHNWTSGRQERPTSARATR--------------ETETASE
MUL_2115|M.ulcerans_Agy99 MTALNDTERAVHNWTSGRQERPTSARATR--------------ETETASE
MAV_3702|M.avium_104 MTALNDSERAVQNWTSARPDRPAPVRSTPPAETA------PKPAAETAVK
Mb2871c|M.bovis_AF2122/97 MTALNDTERAVRNWRAGRPHRPAPMRPPR--------------SEETASE
Rv2846c|M.tuberculosis_H37Rv MTALNDTERAVRNWTAGRPHRPAPMRPPR--------------SEETASE
MLBr_01562|M.leprae_Br4923 MVTLYDHERAVHNWTAARSDRPAPSRLTQ--------------QVEPASE
MSMEG_2619|M.smegmatis_MC2_155 MTALNDAERAAA--RVGLAAR-----------------------------
Mflv_4046|M.gilvum_PYR-GCK MTALNDAERAAIQRDAEGRGKRGSDRASGRALPARVTGSGESAGGRGGDA
TH_2858|M.thermoresistible__bu ---------------MEG--------------------------------
MAB_3142c|M.abscessus_ATCC_199 MSGLS-VEQLTQKAR-----------------------------------
Mvan_2307|M.vanbaalenii_PYR-1 -MASGSSEHATTP-------------------------------------
MMAR_1887|M.marinum_M RASRYYPTWLPSR-----RFIAAVIAIGGMQLLATMDSTVAIVALPKIQD
MUL_2115|M.ulcerans_Agy99 RASRYYPTWLPSR-----RFIAAVIAIGGMQLLATMDSTVAIVALPKIQD
MAV_3702|M.avium_104 RTSKYYPAWLPSR-----RFIAAVIAIGGMQLLATMDSTVAIVALPRIQN
Mb2871c|M.bovis_AF2122/97 RPSRYYPTWLPSR-----SFIAAVIAIGGMQLLATMDSTVAIVALPKIQN
Rv2846c|M.tuberculosis_H37Rv RPSRYYPTWLPSR-----SFIAAVIAIGGMQLLATMDSTVAIVALPKIQN
MLBr_01562|M.leprae_Br4923 RTSK-YPTLLPSGRFPSGRFIAAVIVIGGMQLLATMDSTVAIVALPKIQN
MSMEG_2619|M.smegmatis_MC2_155 -----LPSWFPSWG-----FLSAVIAIGGMQLLATMDSTVAIVALPKIQD
Mflv_4046|M.gilvum_PYR-GCK GRRGRTPAWMPSR-----RFFAAVIAIGGMQLLATMDSTIAIVALPKIQD
TH_2858|M.thermoresistible__bu --ANKLPTWLPSR-----RFIAAVFAIGGMQLLATMDSTIAIVALPRIQD
MAB_3142c|M.abscessus_ATCC_199 -------EFLPSR-----HYLYAVIAIAGMQFLATMDGTIAVVTLPKIQA
Mvan_2307|M.vanbaalenii_PYR-1 ---------LSKS-----VLGIAIVAITGMQLMSTLDGTIVIVALPRMQA
:.. *:..* ***:::*:*.*:.:*:**::*
MMAR_1887|M.marinum_M ELSLSDAGRSWVITAYVLTFGGLMLLGGRLGDTIGRKRTFIVGVALFTIS
MUL_2115|M.ulcerans_Agy99 ELSLSDAGRSWVITAYVLTFGGLMLLGGRLGDTIGRKRTFIVGVALFTIS
MAV_3702|M.avium_104 ELSLSDAGRSWVITAYVLTFGGLMLLGGRLGDTIGRKRTFIVGVALFTIS
Mb2871c|M.bovis_AF2122/97 ELSLSDAGRSWVITAYVLTFGGLMLLGGRLGDTIGRKRTFIVGVALFTIS
Rv2846c|M.tuberculosis_H37Rv ELSLSDAGRSWVITAYVLTFGGLMLLGGRLGDTIGRKRTFIVGVALFTIS
MLBr_01562|M.leprae_Br4923 ELSLSDAGRSWGITAYVLTFGGLMLLGGRLGDTIGRKRTFIVGVALFTIS
MSMEG_2619|M.smegmatis_MC2_155 ELSLSDAGRSWVITAYVLTFGGLMLLGGRLGDTIGRKRTFIVGVMLFTIA
Mflv_4046|M.gilvum_PYR-GCK ELGLSDAGRSWVITAYVLTFGGLMLLGGRLGDTIGRKRTFIVGVALFTIA
TH_2858|M.thermoresistible__bu ELNLTDAGRSWVITAYVLTFGGLMLLGGRLGDTIGRKRTFIVGVTLFTIA
MAB_3142c|M.abscessus_ATCC_199 ELHLNDATRSWVITAYMLSFGGLMLLGGRLGDTFGRKRVFIGGITLFTLA
Mvan_2307|M.vanbaalenii_PYR-1 ELNLSDAGKSWVITAYVLAFGGLLLLGGRIGDAVGHKRAFLSGVGLFTIA
** *.** :** ****:*:****:*****:**:.*:**.*: *: ***::
MMAR_1887|M.marinum_M SVLCAVAWDEATLVIARLSQGVGSAIASPTGLALVATTFPKGPARNAATA
MUL_2115|M.ulcerans_Agy99 SVLCAVAWDEATLVIARLSQGVGSAIASPTGLALVATTFPKGPTRNAATA
MAV_3702|M.avium_104 SVLCAVAWDEATMVIARLSQGVGSAIASPTGLALVATTFPKGPARNFATA
Mb2871c|M.bovis_AF2122/97 SVLCAVAWDEATLVIARLSQGVGSAIASPTGLALVATTFPKGPARNAATA
Rv2846c|M.tuberculosis_H37Rv SVLCAVAWDEATLVIARLSQGVGSAIASPTGLALVATTFPKGPARNAATA
MLBr_01562|M.leprae_Br4923 SVLCAVAWDEVTLVIARLLQGIGSAIASPTGLALVATTFSKGSARNTATA
MSMEG_2619|M.smegmatis_MC2_155 SVLCGIAWNETTLVTARLLQGVGAAIASPTGLALVATTFPKGPARNAATA
Mflv_4046|M.gilvum_PYR-GCK SVLCGIAWDEATLVIARLLQGVGAAIASPTALALIATTFPKGPARNAATA
TH_2858|M.thermoresistible__bu SVLCGIAWDEATLVIARLLQGVGAAIASPTGLALVATTFPKGPARNAATA
MAB_3142c|M.abscessus_ATCC_199 SIGVGLAQEDIGLAIFRLIQGVGAAVASPTALALIATTFPKGPLRTAAVG
Mvan_2307|M.vanbaalenii_PYR-1 SLVCGLATEEITLVAARAFQGAGAAVAAPTGLALIATTYAVGHARNRALA
*: .:* :: :. * ** *:*:*:**.***:***:. * *. * .
MMAR_1887|M.marinum_M VFAAMTAVGSVMGLVVGGALTEVSWRLAFMVNVPIGLVMIYLARTALRET
MUL_2115|M.ulcerans_Agy99 VFAAMTAVGSVMGLVVGGALTEVSWRLAFMVNVPIGLVMIYLARTALRET
MAV_3702|M.avium_104 VFAAMTAVGSVMGLVVGGALTEVSWRLAFLVNVPIGLVMMYLARTALRET
Mb2871c|M.bovis_AF2122/97 VFAAMTAIGSVMGLVVGGALTEVSWRWAFLVNVPIGLVMIYLARTALRET
Rv2846c|M.tuberculosis_H37Rv VFAAMTAIGSVMGLVVGGALTEVSWRWAFLVNVPIGLVMIYLARTALRET
MLBr_01562|M.leprae_Br4923 VFAAMTAIGSVMGLVVGGALTEVSWRLAFLVNVPIGLVMIYLARTALHET
MSMEG_2619|M.smegmatis_MC2_155 VFGAMTAIGSVMGLVVGGALTEVSWRWAFLVNVPIGLVMVYLARTALQET
Mflv_4046|M.gilvum_PYR-GCK VFAAMTGVGSVMGLVVGGALTEVSWRLAFLVNVPIGLIMIYLARKTLRET
TH_2858|M.thermoresistible__bu VFAAMTGVGSVMGLVVGGALTEVSWRLAFLVNVPIGLLVIYLARRTLRET
MAB_3142c|M.abscessus_ATCC_199 VFGAMTGVGSIAGLIVGGALAEVSWRLAFLINVPAGALMIYLAVRSLTET
Mvan_2307|M.vanbaalenii_PYR-1 VSAAMLATGSVLGLVLGGALTVVSWRLAFLINVPIGIAIIAIAVFRLAET
* .** . **: **::****: **** **::*** * :: :* * **
MMAR_1887|M.marinum_M NRERMKLDAAGAILATLACTAAVFAFSMGPEKGWLSITTIGSGAVAFAAA
MUL_2115|M.ulcerans_Agy99 NRERMKLDAAGAILATLACTAAVFAFSMGPEKGWLSITTIGSGAVAFAAA
MAV_3702|M.avium_104 NRERMKLDATGAVLATLACTAAVFAFSMGPEKGWISLTTISSGVVALGAA
Mb2871c|M.bovis_AF2122/97 NKERMKLDATGAILATLACTAAVFAFSIGPEKGWMSGITIGSGLVALAAA
Rv2846c|M.tuberculosis_H37Rv NKERMKLDATGAILATLACTAAVFAFSIGPEKGWMSGITIGSGLVALAAA
MLBr_01562|M.leprae_Br4923 HKERMKLDAAGAILATLACTAAVFAFSMGPEKGWISLTTIGSGLVALVAA
MSMEG_2619|M.smegmatis_MC2_155 NRERMKLDAAGALLATLACTAAVFAFTQGPESGWLAPITLASGAAALVFG
Mflv_4046|M.gilvum_PYR-GCK NRERLKLDAAGALLATMGCTAAVFGFSMGPEAGWMSPVTIGSGIAAVGCL
TH_2858|M.thermoresistible__bu HRERLKLDAAGAILATLGCTAAVFAFSMGPERGWASVITIATGAAALVFL
MAB_3142c|M.abscessus_ATCC_199 EREPMKLDVAGSVLATLGCTAAVFGFTQGPDRGWDSPYTIVSLIAAAALL
Mvan_2307|M.vanbaalenii_PYR-1 HHERLKLDVTGALLATLACTSAVLFFTQGPSRGWADPFVIGAAVASALLF
.:* :***.:*::***:.**:**: *: **. ** .: : .:
MMAR_1887|M.marinum_M LGFVLVERTAENPVVPFDLFHDRNRLVTFIAIFLAGGVMFSLTVCIGLYV
MUL_2115|M.ulcerans_Agy99 LGFVLVERTAENPVVPFDLFHDRNRLVTFIAIFLAGGVMFSLTVCIGLYV
MAV_3702|M.avium_104 LAFVIVERTAENPVVPFDLFRDRNRLVTFIAIFLAGGVMFTLTVCIGLYV
Mb2871c|M.bovis_AF2122/97 VAFVIVERTAENPVVPFHLFRDRNRLVTFSAILLAGGVMFSLTVCIGLYV
Rv2846c|M.tuberculosis_H37Rv VAFVIVERTAENPVVPFHLFRDRNRLVTFSAILLAGGVMFSLTVCIGLYV
MLBr_01562|M.leprae_Br4923 IAFAVVERTAENPVVPFDLFRDRNRLVTFIAIFLAGGVIFSLTVSIGLYI
MSMEG_2619|M.smegmatis_MC2_155 LAFLIAERNAENPVVPFALFRERNRVATFAAIFLAGGVLFTLTVLIGLYV
Mflv_4046|M.gilvum_PYR-GCK IAFLWVERTAENPVVPFELFLDRNRVATFAAVFLAGGVMFTLTVTIGLYV
TH_2858|M.thermoresistible__bu LAFLYVERTADNPVVPFSLFRDRNRVATFGAIFLAGGVMFTLTVLIGLYV
MAB_3142c|M.abscessus_ATCC_199 IAFLFVERTAENPVLPFSLFTDRNRVATFAAIFMAGGVLFTLTITIGLYM
Mvan_2307|M.vanbaalenii_PYR-1 VAFFVVERSADHPIVPLTVFDNRNRVATFLSLFLAGGVMLSATVMIGLYV
:.* .**.*::*::*: :* :***:.** ::::****::: *: ****:
MMAR_1887|M.marinum_M QDVLGYSALRAGVGFIPFVIAMGIGLGVSSQLVSRFSPRVLTIGGGYLLV
MUL_2115|M.ulcerans_Agy99 QDVLGYSALRAGVGFIPFVIAMGIGLGVSSQLVSRFSPRVLTIGGGYLLV
MAV_3702|M.avium_104 QDILGYSALRAGVGFIPFVIAMGIGLGVSSQLVARFSPRVLTIAGGYLLV
Mb2871c|M.bovis_AF2122/97 QDILGYSALRAGVGFIPFVIAMGIGLGVSSQLVSRFSPRVLTIGGGYLLF
Rv2846c|M.tuberculosis_H37Rv QDILGYSALRAGVGFIPFVIAMGIGLGVSSQLVSRFSPRVLTIGGGYLLF
MLBr_01562|M.leprae_Br4923 QDILGYSALRAGIGFIPFVIAMGIGLGVSSQLVSRFSPRVLTIGGGWLLM
MSMEG_2619|M.smegmatis_MC2_155 QDILGYSALRAGVGFIPFVIGMGIGLGAASQLVRSIPPRVLVIAGGILVL
Mflv_4046|M.gilvum_PYR-GCK QDIMGYSALRAGVGFIPFVIALGIGLGVSSALVSKYPPRVLVIGGGVLVL
TH_2858|M.thermoresistible__bu QDIMGYSPLRAGIGFIPFVIALGIGLAASSQLVRLFPPRVLVFGGGVLVL
MAB_3142c|M.abscessus_ATCC_199 QDLMKYSPLRTGIAAIPFVVGMGIGLGLSSQLVTRLAPRVLILCGGVVVF
Mvan_2307|M.vanbaalenii_PYR-1 QDVMGYSALQAGMAFIPFALALGVGTVVAARVAPYLAPRWIILGAGLLVP
**:: **.*::*:. ***.:.:*:* :: :. .** : : .* ::
MMAR_1887|M.marinum_M CAMIYGWAFMQRGVPYFPNLVAPIVIGGIGIGMAVVPLTLAAIAGVGFDR
MUL_2115|M.ulcerans_Agy99 CAMIYGWAFMQRGVPYFPNLVAPIVIGGIGIGMAVVPLTLAAIAGVGFDR
MAV_3702|M.avium_104 LAMLYGWWCMHRGVPYFPNLVLPIVVGGIGIGMAVVPLTLSAIAGVGFDQ
Mb2871c|M.bovis_AF2122/97 GAMLYGSFFMHRGVPYFPNLVMPIVVGGIGIGMAVVPLTLSAIAGVGFDQ
Rv2846c|M.tuberculosis_H37Rv GAMLYGSFFMHRGVPYFPNLVMPIVVGGIGIGMAVVPLTLSAIAGVGFDQ
MLBr_01562|M.leprae_Br4923 VAMLYGWAFMNRGVPYFPNLLAPIVVGGIGIGMAVVPLTLSAIAGVGFDR
MSMEG_2619|M.smegmatis_MC2_155 GAMIYG-STLHRGIPYFPNLVLPITIGGIGIGTIVVPLTLSAIAGVNLDR
Mflv_4046|M.gilvum_PYR-GCK AAMIYG-STLDANIPYFPNLVLPITIGGLGIGMIVVPLMISAIAGVGFDQ
TH_2858|M.thermoresistible__bu GAMLYG-STLHADIPYFPNLVVPIVVGGIGIGVIVVPLTVSAIAGVGFDQ
MAB_3142c|M.abscessus_ATCC_199 GAMLYG-STIDRTIAYFPDFATAIFVGGIGIGTIVVPVILSAITGVDADR
Mvan_2307|M.vanbaalenii_PYR-1 CAMLYG-STLDRGIPYVPDLIAPLVVGGFGIGVISVILPLCAIADVGARE
**:** :. :.*.*:: .: :**:*** * : :.**:.*. .
MMAR_1887|M.marinum_M IGPVSALTLMLQSLGGPLVLAVIQAVITSRTLYLGGTTGPVKFMNDAQLH
MUL_2115|M.ulcerans_Agy99 IGPVSALTLMLQSLGGPLVLAGIQAVITSRTLYLGGTTGPVKFMNDAQLH
MAV_3702|M.avium_104 IGPVSAVTLMLQSLGGPLVLAVIQAVITSRTLYLGGTTGPVKTMNDAQLQ
Mb2871c|M.bovis_AF2122/97 IGPVSAIALMLQSLGGPLVLAVIQAVITSRTLYLGGTTGPVKFMNDVQLA
Rv2846c|M.tuberculosis_H37Rv IGPVSAIALMLQSLGGPLVLAVIQAVITSRTLYLGGTTGPVKFMNDVQLA
MLBr_01562|M.leprae_Br4923 IGPVSAIALMLQSLGGPLVLAVIQAVITSRTLYLGGTTGPVKFMNDAQLR
MSMEG_2619|M.smegmatis_MC2_155 IGPASAIALMLQNLGGPLVLAVIQAVITSRTLFLGGTTGPVKAMNDEQIG
Mflv_4046|M.gilvum_PYR-GCK IGPVSAIALMLQNLGGPVVLAIIQAVITSRTLYLGGTTGPVKDMNAAQMH
TH_2858|M.thermoresistible__bu IGPVSAIALMLQNLGGPLVLAVVQAVITSRTLYLGGTTGPVQNMDTAQLH
MAB_3142c|M.abscessus_ATCC_199 IGPLSAISLMLQNLGGPIVLVIIQAIITSRTLYLGGATGPVQNMNRDQLH
Mvan_2307|M.vanbaalenii_PYR-1 IGPVSSITLMVQNLGGPVVLVVIQAVQTSRTLYLGGTTGPVAAMTPAQLD
*** *:::**:*.****:**. :**: *****:***:**** * *:
MMAR_1887|M.marinum_M ALDHGYTYGLLWVAGAAVIVGGAAVFIGYTPEQVAHAQEVKEAIDAG---
MUL_2115|M.ulcerans_Agy99 ALDHGYTYGLLWVAGAAVIVGGAAVFIGYTPEQVAHAQEVKEAIDAG---
MAV_3702|M.avium_104 ALDHGYTYGLLWVAGAAVIVGAAALFIGYTPEQVAHAQEVKEAMDAG---
Mb2871c|M.bovis_AF2122/97 ALDHAYTYGLLWVAGAAIIVGGMALFIGYTPQQVAHAQEVKEAIDAG---
Rv2846c|M.tuberculosis_H37Rv ALDHAYTYGLLWVAGAAIIVGGMALFIGYTPQQVAHAQEVKEAIDAG---
MLBr_01562|M.leprae_Br4923 ALDHGYTYGLLWVAGAVVIVGVAALFIGYTPEQVAYAQEVKEAVDAG---
MSMEG_2619|M.smegmatis_MC2_155 ALDAAYTYGLLWVAAVAVLVGAAALFIGYTSQQVAHAQDVKDAMDAG---
Mflv_4046|M.gilvum_PYR-GCK ALDQGYSYGLLWVAAVAVIVGAVALFIGYTAQQVAHAQEVKDAIDAG---
TH_2858|M.thermoresistible__bu ALDQGYTYGLLWVAAVAVLVGGAALFIGYTAKQVAHAQEVKDAIDAG---
MAB_3142c|M.abscessus_ATCC_199 ALDHGYTYGMLWIAGTAVLVGVAALFITYSAADIAHAQKAQAAHDQGLDE
Mvan_2307|M.vanbaalenii_PYR-1 ALDAGYTYSLLWVAAVSVLVGAAALFIGFSARQIAVAKYNRDAVETG---
*** .*:*.:**:*.. ::** *:** ::. ::* *: : * : *
MMAR_1887|M.marinum_M -EL------------
MUL_2115|M.ulcerans_Agy99 -EL------------
MAV_3702|M.avium_104 -EL------------
Mb2871c|M.bovis_AF2122/97 -EL------------
Rv2846c|M.tuberculosis_H37Rv -EL------------
MLBr_01562|M.leprae_Br4923 -EL------------
MSMEG_2619|M.smegmatis_MC2_155 -EIEVDQPPIG----
Mflv_4046|M.gilvum_PYR-GCK -EL------------
TH_2858|M.thermoresistible__bu -EL------------
MAB_3142c|M.abscessus_ATCC_199 DELE-----------
Mvan_2307|M.vanbaalenii_PYR-1 -ELWISAEQRGATEG
*: