For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTALNDAERAAARVGLAARLPSWFPSWGFLSAVIAIGGMQLLATMDSTVAIVALPKIQDELSLSDAGRSW VITAYVLTFGGLMLLGGRLGDTIGRKRTFIVGVMLFTIASVLCGIAWNETTLVTARLLQGVGAAIASPTG LALVATTFPKGPARNAATAVFGAMTAIGSVMGLVVGGALTEVSWRWAFLVNVPIGLVMVYLARTALQETN RERMKLDAAGALLATLACTAAVFAFTQGPESGWLAPITLASGAAALVFGLAFLIAERNAENPVVPFALFR ERNRVATFAAIFLAGGVLFTLTVLIGLYVQDILGYSALRAGVGFIPFVIGMGIGLGAASQLVRSIPPRVL VIAGGILVLGAMIYGSTLHRGIPYFPNLVLPITIGGIGIGTIVVPLTLSAIAGVNLDRIGPASAIALMLQ NLGGPLVLAVIQAVITSRTLFLGGTTGPVKAMNDEQIGALDAAYTYGLLWVAAVAVLVGAAALFIGYTSQ QVAHAQDVKDAMDAGEIEVDQPPIG
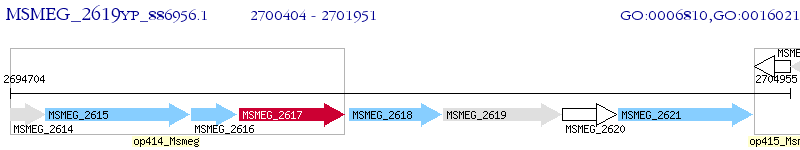
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2619 | - | - | 100% (515) | efflux protein |
| M. smegmatis MC2 155 | MSMEG_5670 | - | 3e-58 | 31.71% (451) | efflux membrane protein |
| M. smegmatis MC2 155 | MSMEG_2848 | - | 6e-46 | 28.98% (459) | transmembrane efflux protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2871c | efpA | 0.0 | 76.98% (530) | integral membrane efflux protein EfpA |
| M. gilvum PYR-GCK | Mflv_4046 | - | 0.0 | 76.06% (543) | EmrB/QacA family drug resistance transporter |
| M. tuberculosis H37Rv | Rv2846c | efpA | 0.0 | 76.98% (530) | integral membrane efflux protein EfpA |
| M. leprae Br4923 | MLBr_01562 | - | 0.0 | 78.57% (490) | putative transmembrane efflux protein |
| M. abscessus ATCC 19977 | MAB_3142c | - | 0.0 | 65.51% (490) | putative transmembrane efflux protein |
| M. marinum M | MMAR_1887 | efpA | 0.0 | 75.47% (530) | integral membrane efflux protein EfpA |
| M. avium 104 | MAV_3702 | - | 0.0 | 80.74% (488) | transporter, major facilitator family protein |
| M. thermoresistible (build 8) | TH_2858 | - | 0.0 | 81.87% (491) | PUTATIVE major facilitator superfamily MFS_1 |
| M. ulcerans Agy99 | MUL_2115 | efpA | 0.0 | 75.09% (530) | integral membrane efflux protein EfpA |
| M. vanbaalenii PYR-1 | Mvan_2306 | - | 0.0 | 76.80% (543) | major facilitator superfamily transporter |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4046|M.gilvum_PYR-GCK MTALNDAERAAIQRDAEGRGKRGSDRASGRALPARVTGSGESAGGRGGDA
Mvan_2306|M.vanbaalenii_PYR-1 MTALNDAERAAIQRDAEGSGKRGSGRASDRALPARIPGLGETAGGRGGDG
TH_2858|M.thermoresistible__bu ---------------MEG--------------------------------
MMAR_1887|M.marinum_M MTALNDTERAVHNWTSGRQERPTSARATR--------------ETETASE
MUL_2115|M.ulcerans_Agy99 MTALNDTERAVHNWTSGRQERPTSARATR--------------ETETASE
MAV_3702|M.avium_104 MTALNDSERAVQNWTSARPDRPAPVRSTPPAETAPKP------AAETAVK
Mb2871c|M.bovis_AF2122/97 MTALNDTERAVRNWRAGRPHRPAPMRPPR--------------SEETASE
Rv2846c|M.tuberculosis_H37Rv MTALNDTERAVRNWTAGRPHRPAPMRPPR--------------SEETASE
MLBr_01562|M.leprae_Br4923 MVTLYDHERAVHNWTAARSDRPAPSRLTQ--------------QVEPASE
MSMEG_2619|M.smegmatis_MC2_155 MTALNDAERAAA--RVGLAAR-----------------------------
MAB_3142c|M.abscessus_ATCC_199 MSGLS-------------------------------------------VE
Mflv_4046|M.gilvum_PYR-GCK GRRGRTPAWMPSR-----RFFAAVIAIGGMQLLATMDSTIAIVALPKIQD
Mvan_2306|M.vanbaalenii_PYR-1 ARAGRTPAWVPSR-----RFFAAVIAIGGMQLLATMDSTIAIVALPKIQD
TH_2858|M.thermoresistible__bu --ANKLPTWLPSR-----RFIAAVFAIGGMQLLATMDSTIAIVALPRIQD
MMAR_1887|M.marinum_M RASRYYPTWLPSR-----RFIAAVIAIGGMQLLATMDSTVAIVALPKIQD
MUL_2115|M.ulcerans_Agy99 RASRYYPTWLPSR-----RFIAAVIAIGGMQLLATMDSTVAIVALPKIQD
MAV_3702|M.avium_104 RTSKYYPAWLPSR-----RFIAAVIAIGGMQLLATMDSTVAIVALPRIQN
Mb2871c|M.bovis_AF2122/97 RPSRYYPTWLPSR-----SFIAAVIAIGGMQLLATMDSTVAIVALPKIQN
Rv2846c|M.tuberculosis_H37Rv RPSRYYPTWLPSR-----SFIAAVIAIGGMQLLATMDSTVAIVALPKIQN
MLBr_01562|M.leprae_Br4923 RTSK-YPTLLPSGRFPSGRFIAAVIVIGGMQLLATMDSTVAIVALPKIQN
MSMEG_2619|M.smegmatis_MC2_155 -----LPSWFPSWG-----FLSAVIAIGGMQLLATMDSTVAIVALPKIQD
MAB_3142c|M.abscessus_ATCC_199 QLTQKAREFLPSR-----HYLYAVIAIAGMQFLATMDGTIAVVTLPKIQA
.** :: **:.*.***:*****.*:*:*:**:**
Mflv_4046|M.gilvum_PYR-GCK ELGLSDAGRSWVITAYVLTFGGLMLLGGRLGDTIGRKRTFIVGVALFTIA
Mvan_2306|M.vanbaalenii_PYR-1 ELNLSDAGRSWVITAYVLTFGGLMLLGGRLGDTIGRKRTFIVGVTLFTIA
TH_2858|M.thermoresistible__bu ELNLTDAGRSWVITAYVLTFGGLMLLGGRLGDTIGRKRTFIVGVTLFTIA
MMAR_1887|M.marinum_M ELSLSDAGRSWVITAYVLTFGGLMLLGGRLGDTIGRKRTFIVGVALFTIS
MUL_2115|M.ulcerans_Agy99 ELSLSDAGRSWVITAYVLTFGGLMLLGGRLGDTIGRKRTFIVGVALFTIS
MAV_3702|M.avium_104 ELSLSDAGRSWVITAYVLTFGGLMLLGGRLGDTIGRKRTFIVGVALFTIS
Mb2871c|M.bovis_AF2122/97 ELSLSDAGRSWVITAYVLTFGGLMLLGGRLGDTIGRKRTFIVGVALFTIS
Rv2846c|M.tuberculosis_H37Rv ELSLSDAGRSWVITAYVLTFGGLMLLGGRLGDTIGRKRTFIVGVALFTIS
MLBr_01562|M.leprae_Br4923 ELSLSDAGRSWGITAYVLTFGGLMLLGGRLGDTIGRKRTFIVGVALFTIS
MSMEG_2619|M.smegmatis_MC2_155 ELSLSDAGRSWVITAYVLTFGGLMLLGGRLGDTIGRKRTFIVGVMLFTIA
MAB_3142c|M.abscessus_ATCC_199 ELHLNDATRSWVITAYMLSFGGLMLLGGRLGDTFGRKRVFIGGITLFTLA
** *.** *** ****:*:**************:****.** *: ***::
Mflv_4046|M.gilvum_PYR-GCK SVLCGIAWDEATLVIARLLQGVGAAIASPTALALIATTFPKGPARNAATA
Mvan_2306|M.vanbaalenii_PYR-1 SVLCGIAWDEATLVIARLLQGVGAAIASPTALALIATTFPKGPARNAATA
TH_2858|M.thermoresistible__bu SVLCGIAWDEATLVIARLLQGVGAAIASPTGLALVATTFPKGPARNAATA
MMAR_1887|M.marinum_M SVLCAVAWDEATLVIARLSQGVGSAIASPTGLALVATTFPKGPARNAATA
MUL_2115|M.ulcerans_Agy99 SVLCAVAWDEATLVIARLSQGVGSAIASPTGLALVATTFPKGPTRNAATA
MAV_3702|M.avium_104 SVLCAVAWDEATMVIARLSQGVGSAIASPTGLALVATTFPKGPARNFATA
Mb2871c|M.bovis_AF2122/97 SVLCAVAWDEATLVIARLSQGVGSAIASPTGLALVATTFPKGPARNAATA
Rv2846c|M.tuberculosis_H37Rv SVLCAVAWDEATLVIARLSQGVGSAIASPTGLALVATTFPKGPARNAATA
MLBr_01562|M.leprae_Br4923 SVLCAVAWDEVTLVIARLLQGIGSAIASPTGLALVATTFSKGSARNTATA
MSMEG_2619|M.smegmatis_MC2_155 SVLCGIAWNETTLVTARLLQGVGAAIASPTGLALVATTFPKGPARNAATA
MAB_3142c|M.abscessus_ATCC_199 SIGVGLAQEDIGLAIFRLIQGVGAAVASPTALALIATTFPKGPLRTAAVG
*: .:* :: :. ** **:*:*:****.***:****.**. *. *..
Mflv_4046|M.gilvum_PYR-GCK VFAAMTGVGSVMGLVVGGALTEVSWRLAFLVNVPIGLIMIYLARKTLRET
Mvan_2306|M.vanbaalenii_PYR-1 VFAAMTGVGSVMGLVVGGALTEVSWRLAFLVNVPIGLIMIYLARKTLRET
TH_2858|M.thermoresistible__bu VFAAMTGVGSVMGLVVGGALTEVSWRLAFLVNVPIGLLVIYLARRTLRET
MMAR_1887|M.marinum_M VFAAMTAVGSVMGLVVGGALTEVSWRLAFMVNVPIGLVMIYLARTALRET
MUL_2115|M.ulcerans_Agy99 VFAAMTAVGSVMGLVVGGALTEVSWRLAFMVNVPIGLVMIYLARTALRET
MAV_3702|M.avium_104 VFAAMTAVGSVMGLVVGGALTEVSWRLAFLVNVPIGLVMMYLARTALRET
Mb2871c|M.bovis_AF2122/97 VFAAMTAIGSVMGLVVGGALTEVSWRWAFLVNVPIGLVMIYLARTALRET
Rv2846c|M.tuberculosis_H37Rv VFAAMTAIGSVMGLVVGGALTEVSWRWAFLVNVPIGLVMIYLARTALRET
MLBr_01562|M.leprae_Br4923 VFAAMTAIGSVMGLVVGGALTEVSWRLAFLVNVPIGLVMIYLARTALHET
MSMEG_2619|M.smegmatis_MC2_155 VFGAMTAIGSVMGLVVGGALTEVSWRWAFLVNVPIGLVMVYLARTALQET
MAB_3142c|M.abscessus_ATCC_199 VFGAMTGVGSIAGLIVGGALAEVSWRLAFLINVPAGALMIYLAVRSLTET
**.***.:**: **:*****:***** **::*** * :::*** :* **
Mflv_4046|M.gilvum_PYR-GCK NRERLKLDAAGALLATMGCTAAVFGFSMGPEAGWMSPVTIGSGIAAVGCL
Mvan_2306|M.vanbaalenii_PYR-1 NKERLKLDAAGALLATMGCTAAVFAFSMGPESGWASPVTVASAVAAVGCL
TH_2858|M.thermoresistible__bu HRERLKLDAAGAILATLGCTAAVFAFSMGPERGWASVITIATGAAALVFL
MMAR_1887|M.marinum_M NRERMKLDAAGAILATLACTAAVFAFSMGPEKGWLSITTIGSGAVAFAAA
MUL_2115|M.ulcerans_Agy99 NRERMKLDAAGAILATLACTAAVFAFSMGPEKGWLSITTIGSGAVAFAAA
MAV_3702|M.avium_104 NRERMKLDATGAVLATLACTAAVFAFSMGPEKGWISLTTISSGVVALGAA
Mb2871c|M.bovis_AF2122/97 NKERMKLDATGAILATLACTAAVFAFSIGPEKGWMSGITIGSGLVALAAA
Rv2846c|M.tuberculosis_H37Rv NKERMKLDATGAILATLACTAAVFAFSIGPEKGWMSGITIGSGLVALAAA
MLBr_01562|M.leprae_Br4923 HKERMKLDAAGAILATLACTAAVFAFSMGPEKGWISLTTIGSGLVALVAA
MSMEG_2619|M.smegmatis_MC2_155 NRERMKLDAAGALLATLACTAAVFAFTQGPESGWLAPITLASGAAALVFG
MAB_3142c|M.abscessus_ATCC_199 EREPMKLDVAGSVLATLGCTAAVFGFTQGPDRGWDSPYTIVSLIAAAALL
.:* :***.:*::***:.******.*: **: ** : *: : .*
Mflv_4046|M.gilvum_PYR-GCK IAFLWVERTAENPVVPFELFLDRNRVATFAAVFLAGGVMFTLTVTIGLYV
Mvan_2306|M.vanbaalenii_PYR-1 LAFLWVERTAENPVVPFALFHDRNRVATFAAVFLAGGVMFTLTVTIGLYV
TH_2858|M.thermoresistible__bu LAFLYVERTADNPVVPFSLFRDRNRVATFGAIFLAGGVMFTLTVLIGLYV
MMAR_1887|M.marinum_M LGFVLVERTAENPVVPFDLFHDRNRLVTFIAIFLAGGVMFSLTVCIGLYV
MUL_2115|M.ulcerans_Agy99 LGFVLVERTAENPVVPFDLFHDRNRLVTFIAIFLAGGVMFSLTVCIGLYV
MAV_3702|M.avium_104 LAFVIVERTAENPVVPFDLFRDRNRLVTFIAIFLAGGVMFTLTVCIGLYV
Mb2871c|M.bovis_AF2122/97 VAFVIVERTAENPVVPFHLFRDRNRLVTFSAILLAGGVMFSLTVCIGLYV
Rv2846c|M.tuberculosis_H37Rv VAFVIVERTAENPVVPFHLFRDRNRLVTFSAILLAGGVMFSLTVCIGLYV
MLBr_01562|M.leprae_Br4923 IAFAVVERTAENPVVPFDLFRDRNRLVTFIAIFLAGGVIFSLTVSIGLYI
MSMEG_2619|M.smegmatis_MC2_155 LAFLIAERNAENPVVPFALFRERNRVATFAAIFLAGGVLFTLTVLIGLYV
MAB_3142c|M.abscessus_ATCC_199 IAFLFVERTAENPVLPFSLFTDRNRVATFAAIFMAGGVLFTLTITIGLYM
:.* .**.*:***:** ** :***:.** *:::****:*:**: ****:
Mflv_4046|M.gilvum_PYR-GCK QDIMGYSALRAGVGFIPFVIALGIGLGVSSALVSKYPPRVLVIGGGVLVL
Mvan_2306|M.vanbaalenii_PYR-1 QDIMGYSALRAGIGFIPFVIALGIGLGLSSALVSKFPPRVLVIGGGVLVL
TH_2858|M.thermoresistible__bu QDIMGYSPLRAGIGFIPFVIALGIGLAASSQLVRLFPPRVLVFGGGVLVL
MMAR_1887|M.marinum_M QDVLGYSALRAGVGFIPFVIAMGIGLGVSSQLVSRFSPRVLTIGGGYLLV
MUL_2115|M.ulcerans_Agy99 QDVLGYSALRAGVGFIPFVIAMGIGLGVSSQLVSRFSPRVLTIGGGYLLV
MAV_3702|M.avium_104 QDILGYSALRAGVGFIPFVIAMGIGLGVSSQLVARFSPRVLTIAGGYLLV
Mb2871c|M.bovis_AF2122/97 QDILGYSALRAGVGFIPFVIAMGIGLGVSSQLVSRFSPRVLTIGGGYLLF
Rv2846c|M.tuberculosis_H37Rv QDILGYSALRAGVGFIPFVIAMGIGLGVSSQLVSRFSPRVLTIGGGYLLF
MLBr_01562|M.leprae_Br4923 QDILGYSALRAGIGFIPFVIAMGIGLGVSSQLVSRFSPRVLTIGGGWLLM
MSMEG_2619|M.smegmatis_MC2_155 QDILGYSALRAGVGFIPFVIGMGIGLGAASQLVRSIPPRVLVIAGGILVL
MAB_3142c|M.abscessus_ATCC_199 QDLMKYSPLRTGIAAIPFVVGMGIGLGLSSQLVTRLAPRVLILCGGVVVF
**:: **.**:*:. ****:.:****. :* ** .**** : ** ::.
Mflv_4046|M.gilvum_PYR-GCK AAMIYG-STLDANIPYFPNLVLPITIGGLGIGMIVVPLMISAIAGVGFDQ
Mvan_2306|M.vanbaalenii_PYR-1 AAMIYG-STLDAGIPYFPNLVLPITIGGFGIGMIVVPLMISAIAGVGFDQ
TH_2858|M.thermoresistible__bu GAMLYG-STLHADIPYFPNLVVPIVVGGIGIGVIVVPLTVSAIAGVGFDQ
MMAR_1887|M.marinum_M CAMIYGWAFMQRGVPYFPNLVAPIVIGGIGIGMAVVPLTLAAIAGVGFDR
MUL_2115|M.ulcerans_Agy99 CAMIYGWAFMQRGVPYFPNLVAPIVIGGIGIGMAVVPLTLAAIAGVGFDR
MAV_3702|M.avium_104 LAMLYGWWCMHRGVPYFPNLVLPIVVGGIGIGMAVVPLTLSAIAGVGFDQ
Mb2871c|M.bovis_AF2122/97 GAMLYGSFFMHRGVPYFPNLVMPIVVGGIGIGMAVVPLTLSAIAGVGFDQ
Rv2846c|M.tuberculosis_H37Rv GAMLYGSFFMHRGVPYFPNLVMPIVVGGIGIGMAVVPLTLSAIAGVGFDQ
MLBr_01562|M.leprae_Br4923 VAMLYGWAFMNRGVPYFPNLLAPIVVGGIGIGMAVVPLTLSAIAGVGFDR
MSMEG_2619|M.smegmatis_MC2_155 GAMIYG-STLHRGIPYFPNLVLPITIGGIGIGTIVVPLTLSAIAGVNLDR
MAB_3142c|M.abscessus_ATCC_199 GAMLYG-STIDRTIAYFPDFATAIFVGGIGIGTIVVPVILSAITGVDADR
**:** :. :.***:: .* :**:*** ***: ::**:**. *:
Mflv_4046|M.gilvum_PYR-GCK IGPVSAIALMLQNLGGPVVLAIIQAVITSRTLYLGGTTGPVKDMNAAQMH
Mvan_2306|M.vanbaalenii_PYR-1 IGPVSAIALMLQNLGGPVVLAVIQAVITSRTLYLGGTTGPVKDMNAAQLH
TH_2858|M.thermoresistible__bu IGPVSAIALMLQNLGGPLVLAVVQAVITSRTLYLGGTTGPVQNMDTAQLH
MMAR_1887|M.marinum_M IGPVSALTLMLQSLGGPLVLAVIQAVITSRTLYLGGTTGPVKFMNDAQLH
MUL_2115|M.ulcerans_Agy99 IGPVSALTLMLQSLGGPLVLAGIQAVITSRTLYLGGTTGPVKFMNDAQLH
MAV_3702|M.avium_104 IGPVSAVTLMLQSLGGPLVLAVIQAVITSRTLYLGGTTGPVKTMNDAQLQ
Mb2871c|M.bovis_AF2122/97 IGPVSAIALMLQSLGGPLVLAVIQAVITSRTLYLGGTTGPVKFMNDVQLA
Rv2846c|M.tuberculosis_H37Rv IGPVSAIALMLQSLGGPLVLAVIQAVITSRTLYLGGTTGPVKFMNDVQLA
MLBr_01562|M.leprae_Br4923 IGPVSAIALMLQSLGGPLVLAVIQAVITSRTLYLGGTTGPVKFMNDAQLR
MSMEG_2619|M.smegmatis_MC2_155 IGPASAIALMLQNLGGPLVLAVIQAVITSRTLFLGGTTGPVKAMNDEQIG
MAB_3142c|M.abscessus_ATCC_199 IGPLSAISLMLQNLGGPIVLVIIQAIITSRTLYLGGATGPVQNMNRDQLH
*** **::****.****:**. :**:******:***:****: *: *:
Mflv_4046|M.gilvum_PYR-GCK ALDQGYSYGLLWVAAVAVIVGAVALFIGYTAQQVAHAQEVKDAIDAG---
Mvan_2306|M.vanbaalenii_PYR-1 ALDQGYSYGLLWVAAVAVIVGGAALFIGYTAEQVAHAQEVKDAIDAG---
TH_2858|M.thermoresistible__bu ALDQGYTYGLLWVAAVAVLVGGAALFIGYTAKQVAHAQEVKDAIDAG---
MMAR_1887|M.marinum_M ALDHGYTYGLLWVAGAAVIVGGAAVFIGYTPEQVAHAQEVKEAIDAG---
MUL_2115|M.ulcerans_Agy99 ALDHGYTYGLLWVAGAAVIVGGAAVFIGYTPEQVAHAQEVKEAIDAG---
MAV_3702|M.avium_104 ALDHGYTYGLLWVAGAAVIVGAAALFIGYTPEQVAHAQEVKEAMDAG---
Mb2871c|M.bovis_AF2122/97 ALDHAYTYGLLWVAGAAIIVGGMALFIGYTPQQVAHAQEVKEAIDAG---
Rv2846c|M.tuberculosis_H37Rv ALDHAYTYGLLWVAGAAIIVGGMALFIGYTPQQVAHAQEVKEAIDAG---
MLBr_01562|M.leprae_Br4923 ALDHGYTYGLLWVAGAVVIVGVAALFIGYTPEQVAYAQEVKEAVDAG---
MSMEG_2619|M.smegmatis_MC2_155 ALDAAYTYGLLWVAAVAVLVGAAALFIGYTSQQVAHAQDVKDAMDAG---
MAB_3142c|M.abscessus_ATCC_199 ALDHGYTYGMLWIAGTAVLVGVAALFITYSAADIAHAQKAQAAHDQGLDE
*** .*:**:**:*...::** *:** *:. ::*:**..: * * *
Mflv_4046|M.gilvum_PYR-GCK -EL--------
Mvan_2306|M.vanbaalenii_PYR-1 -EL--------
TH_2858|M.thermoresistible__bu -EL--------
MMAR_1887|M.marinum_M -EL--------
MUL_2115|M.ulcerans_Agy99 -EL--------
MAV_3702|M.avium_104 -EL--------
Mb2871c|M.bovis_AF2122/97 -EL--------
Rv2846c|M.tuberculosis_H37Rv -EL--------
MLBr_01562|M.leprae_Br4923 -EL--------
MSMEG_2619|M.smegmatis_MC2_155 -EIEVDQPPIG
MAB_3142c|M.abscessus_ATCC_199 DELE-------
*: