For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MASQVLQDKVVFVTGAARGQGRAHAVRFAEEGADVIAFDLCAQIDSVAYPMATPEDLDETVRLVEKTGRR IVAERGDVRDRDRLAEVVARGVAEFGRLDFVLANAGILPAAGPQGLDITAFTDAIAVMLNGVYFTIEAAL PALLRNPDGGAIVITSSAAGFTSVSTEFGTMSHGAAGYTAAKHGVIGVMRHFARSLAEKNIRVNSVHPGG VATPMVLNQAMADWSGEHPSFSAAQQPLLRLPMMEPEAVSDAMVYLCGPSGRYLTGATLPLDAGQTLK
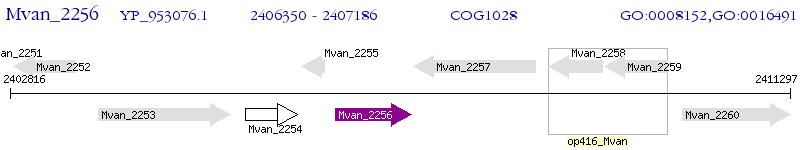
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_2256 | - | - | 100% (278) | short-chain dehydrogenase/reductase SDR |
| M. vanbaalenii PYR-1 | Mvan_2391 | - | 3e-65 | 49.08% (273) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. vanbaalenii PYR-1 | Mvan_3394 | - | 1e-60 | 44.85% (272) | short chain dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2771 | - | 9e-64 | 48.19% (276) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. gilvum PYR-GCK | Mflv_4087 | - | 1e-133 | 85.93% (270) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv2750 | - | 1e-63 | 48.19% (276) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. leprae Br4923 | MLBr_01740 | - | 4e-17 | 30.74% (231) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3081 | - | 6e-64 | 48.36% (275) | Short-chain dehydrogenase/reductase |
| M. marinum M | MMAR_1965 | fabG | 7e-63 | 46.21% (277) | short chain dehydrogenase |
| M. avium 104 | MAV_3641 | - | 4e-69 | 50.92% (273) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. smegmatis MC2 155 | MSMEG_2687 | - | 2e-67 | 50.55% (273) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. thermoresistible (build 8) | TH_0123 | - | 3e-71 | 51.85% (270) | PROBABLE DEHYDROGENASE |
| M. ulcerans Agy99 | MUL_3392 | fabG | 2e-62 | 45.85% (277) | 3-ketoacyl-(acyl-carrier-protein) reductase |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_2256|M.vanbaalenii_PYR-1 ----MASQVLQDKVVFVTGAARGQGRAHAVRFAEEGADVIAFDLCA-QID
Mflv_4087|M.gilvum_PYR-GCK MTADAPHGLFDGKVVLVTGAARGQGRAHAVRFAEEGADVIAVDVCA-QLD
Mb2771|M.bovis_AF2122/97 ---MIDR-PLEGKVAFITGAARGLGRAHAVRLAADGANIIAVDICE-QIA
Rv2750|M.tuberculosis_H37Rv ---MIDR-PLEGKVAFITGAARGLGRAHAVRLAADGANIIAVDICE-QIA
MSMEG_2687|M.smegmatis_MC2_155 ---MSDTAPLSGKVAFITGAARGQGRAEAVRLAADGADIIAVDLCA-QIA
MAV_3641|M.avium_104 -MPRSSEGPLTGKVAFITGAARGQGRAHAVRLAADGADIIAVDLCD-QIA
MMAR_1965|M.marinum_M -MSRPLDGPLSGRVAFITGAARGQGRAHALRLARDGADVIAVDLCD-QIA
MUL_3392|M.ulcerans_Agy99 -MSRPLDGPLSGRVAFITGAARGQGRAHALRLARDGADVIAVDLCD-QIA
MAB_3081|M.abscessus_ATCC_1997 ------MGKLDGKVAFITGAARGQGRAHAIKLASEGASIIAVDICA-QIP
TH_0123|M.thermoresistible__bu ------MGRVQGKVAFITGAARGQGRSHAVRLAEEGASIIAVDSCQGSMD
MLBr_01740|M.leprae_Br4923 --MVKTAQYFVGKRCFVTGAASGIGRATALRLAAYGAELYLTDRHC----
. .: ::**** * **: *:::* **.: *
Mvan_2256|M.vanbaalenii_PYR-1 SVAYPMATPEDLDETVRLVEKTGRRIVAERG-DVRDRDRLAEVVARGVAE
Mflv_4087|M.gilvum_PYR-GCK SVAYPMSTPEDLDETARLVEKTGRRVVAEQC-DVRDRDRLADVVAAGVEE
Mb2771|M.bovis_AF2122/97 SVPYPLSTADDLAATVELVEDAGGGIVARQG-DVRDRASLSVALQAGLDE
Rv2750|M.tuberculosis_H37Rv SVPYPLSTADDLAATVELVEDAGGGIVARQG-DVRDRASLSVALQAGLDE
MSMEG_2687|M.smegmatis_MC2_155 SVPYPLATPDDLATTAELVEKAGGRIVTRQA-DVRDEPELAAALAAGLDE
MAV_3641|M.avium_104 SVPYPLATPEELAATVKLVEDIGSRIVARQA-DVRDRESLSAALQAGLDE
MMAR_1965|M.marinum_M SVPYPLGTAEELATTVKLVEDTGARIVASQA-DVRDREALAAALQAGIDE
MUL_3392|M.ulcerans_Agy99 SVPYPLGTAEELATTVKLVEDTGARIVASQA-DVRDREALAAALQAGIDE
MAB_3081|M.abscessus_ATCC_1997 SVEYPMATTEDLDLTVKEVDALGRAISAHET-DVRDREALQAAFDAGVHD
TH_0123|M.thermoresistible__bu TLLYDLPTEDDLKETCNQVEAVGGRIVATKA-DVRDRKGLRDAFEAGYAE
MLBr_01740|M.leprae_Br4923 ---------DGLAQTVSEARALGAQVPEYRALDISDYDEVAAFGADIHAR
: * * . * : . *: * :
Mvan_2256|M.vanbaalenii_PYR-1 FGRLDFVLANAGILPAAGPQGLDITAFTDAIAVMLNGVYFTIEAALPALL
Mflv_4087|M.gilvum_PYR-GCK FGRLDFVLANAGILPIAGDQGRSITAFTDAVAVMLNGVYFTVDAALPALL
Mb2771|M.bovis_AF2122/97 FGRLDIVVANAGIAMMQ----AGDDGWRDVIDVNLTGVFHTVQVAIPTLI
Rv2750|M.tuberculosis_H37Rv FGRLDIVVANAGIAMMQ----AGDDGWRDVIDVNLTGVFHTVQVAIPTLI
MSMEG_2687|M.smegmatis_MC2_155 FGRLDIVVANAGIAPML----SGPEGWRDVIDVNLTGVHHTVEVAIPTLV
MAV_3641|M.avium_104 FGRLDIVVANAGIAPMS----AGDDGWHDVIDVNLTGVYHTIKVAIPALV
MMAR_1965|M.marinum_M LGQVDIVVANAGIAPMQ----SGDDGWRDVIDVNLSGAYYTVEVAIPTMI
MUL_3392|M.ulcerans_Agy99 LGQVDIVVANAGIAPMQ----SGDDGWRDVIDVNLSGAYYTVEVAIPTMI
MAB_3081|M.abscessus_ATCC_1997 LGPVDIVVANAGIATLTPE--CGDDGWRDVLDVNLTGVYHTIEVARPSMV
TH_0123|M.thermoresistible__bu FGRVDIVLGNAGIMPALGEVSDTDEAFDDAIAVMLTGVYNTLRVATPKLV
MLBr_01740|M.leprae_Br4923 HPSMDIVMNIAGVSVWGTVDRLTHDQWSNVVAINLMGPIHIIETFVPAIL
:*:*: **: : :.: : * * : . * ::
Mvan_2256|M.vanbaalenii_PYR-1 RNPDGGAIVITSSAAGFTSVSTEFGTMSHGAAGYTAAKHGVIGVMRHFAR
Mflv_4087|M.gilvum_PYR-GCK RNPDGGAIVITSSAAGFTSVSTGFDTMNHGAAGYTAAKHGVIGVMRHFAR
Mb2771|M.bovis_AF2122/97 EQGTGGSIVLISSAAGLVGIGSS----DPGSLGYAAAKHGVVGLMRAYAN
Rv2750|M.tuberculosis_H37Rv EQGTGGSIVLISSAAGLVGIGSS----DPGSLGYAAAKHGVVGLMRAYAN
MSMEG_2687|M.smegmatis_MC2_155 EQGDGGAIVLISSVAGLVGIGGG----DRGSLGYTAAKHGIVGLMRAYAN
MAV_3641|M.avium_104 KQGTGGSIVLISSSAGLAGVGSA----DPGSVGYVAAKHGVVGLMRVYAN
MMAR_1965|M.marinum_M EQGRGGSIVLISSAAGLVGISSA----DAGAIGYAASKHALVGLMRVYAN
MUL_3392|M.ulcerans_Agy99 EQGRGGSIVLISSAAGLVGISSA----DAGAIGYVASKHALVGLMRVYAN
MAB_3081|M.abscessus_ATCC_1997 QRGAGGSIVLTSSVAGLTGMVMD----NPGGLAYTAAKHGLVGLMRSYAN
TH_0123|M.thermoresistible__bu EQGQGGSIVLTSSTAGLKGQTTG----TAGCDGYVAAKHGVVGLMRAYAT
MLBr_01740|M.leprae_Br4923 AAGRGGHLVNVSSAAGLVALPWH--------AAYSASKYGLRGLSEVLRF
** :* ** **: . .* *:*:.: *: .
Mvan_2256|M.vanbaalenii_PYR-1 SLAEKNIRVNSVHPGGVATPMVL----------NQAMADWSG---EHPSF
Mflv_4087|M.gilvum_PYR-GCK SLAEKNIRVNSVHPGGVATPMVL----------NEALAQWAG---EHPSF
Mb2771|M.bovis_AF2122/97 HLAPQNIRVNSVHPCGVDTPMIN----------NEFFQQWLT--TADMDA
Rv2750|M.tuberculosis_H37Rv HLAPQNIRVNSVHPCGVDTPMIN----------NEFFQQWLT--TADMDA
MSMEG_2687|M.smegmatis_MC2_155 HLAPHSIRVNSVHPTGVDTPMID----------NEFTRGWLAHITEETGR
MAV_3641|M.avium_104 LLAGQMIRVNSIHPSGVETPMIN----------NEFTREWLAKMAAATDT
MMAR_1965|M.marinum_M LLAPHSIRVNSLHPSGVDTPMIN----------NEFIRRWLADLVAETGS
MUL_3392|M.ulcerans_Agy99 LLAPHSIRVNSLHPSGVDPPMIN----------NEFIRHWLADLVAETGS
MAB_3081|M.abscessus_ATCC_1997 MLAPHNIRVNTIHPSGVATPMLI----------NPVTEKLFADGSPEGMS
TH_0123|M.thermoresistible__bu TYAKHNIRVNTVHPCGCATPMIE----------NVAFAQYAQ---ENPET
MLBr_01740|M.leprae_Br4923 DLARYRIGVSVVVPGAVRTPLVDTIEIAGVDRQDPRFSRWVDRFRGHAVS
* * *. : * . .*:: :
Mvan_2256|M.vanbaalenii_PYR-1 SAAQQPLLRLPMME---------------------------PEAVSDAMV
Mflv_4087|M.gilvum_PYR-GCK SQAQQALLAMPMME---------------------------PEHISDTMV
Mb2771|M.bovis_AF2122/97 PHNLGNALPVELVQ---------------------------PTDIANAVA
Rv2750|M.tuberculosis_H37Rv PHNLGNALPVELVQ---------------------------PTDIANAVA
MSMEG_2687|M.smegmatis_MC2_155 PVDMGNALPVQAID---------------------------ADDVANAVA
MAV_3641|M.avium_104 PGAMGNAMPVEVLA---------------------------PEDVANAVA
MMAR_1965|M.marinum_M GPGAGNALPVQILQ---------------------------ADDIAGALA
MUL_3392|M.ulcerans_Agy99 GPGAGNALPVQILQ---------------------------ADDIAGALA
MAB_3081|M.abscessus_ATCC_1997 --DTGNALPVVLME---------------------------PEDMANTVA
TH_0123|M.thermoresistible__bu VSTWQTMLPVPQVQ---------------------------PMDISNAIL
MLBr_01740|M.leprae_Br4923 AERAAEKILAGVAKNRYLIYTSHDIRVLYGFKRLAWWPYGVVMKQVNVVF
: ..:
Mvan_2256|M.vanbaalenii_PYR-1 YLCGPSGRYLTGATLPLDAGQTLK-----------
Mflv_4087|M.gilvum_PYR-GCK YLCGPAGRYVTGVALPVDAGQTLK-----------
Mb2771|M.bovis_AF2122/97 WLASEEARYVTGVTLPVDAGFVNKR----------
Rv2750|M.tuberculosis_H37Rv WLASEEARYVTGVTLPVDAGFVNKR----------
MSMEG_2687|M.smegmatis_MC2_155 WLVSDQARYVTGVTLPVDAGIVNKR----------
MAV_3641|M.avium_104 WLVSDQARYITGVTLPVDAGFLNK-----------
MMAR_1965|M.marinum_M WLVSDEARYITGVALPVDAGCVNKR----------
MUL_3392|M.ulcerans_Agy99 WLVSDEARYITGVALPVDAGCVNKR----------
MAB_3081|M.abscessus_ATCC_1997 WLVSEDARYITGVALPVDAGFTNRR----------
TH_0123|M.thermoresistible__bu WLCSDEARYVTGVTLPVDAGAVAR-----------
MLBr_01740|M.leprae_Br4923 TRALRPSPATMRCVEPIKVGVYSQPRSGEGGKSGN
. . *:..* :