For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MRLVYLIAVPVVVSTGIAIAPPAVADCTSAGGTTICSQGEVRGSTGGGGPSGPYVPYPCDLNYYECNEWW DFDADIDIDPGPPGIGAPGRPDIGGPGRPGGGRPGGGGGGIGGGRGR
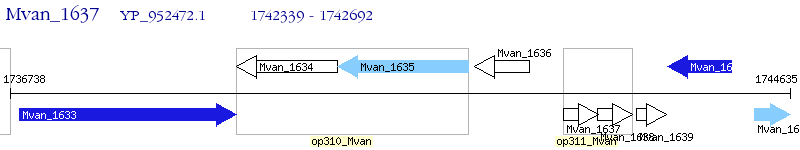
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_1637 | - | - | 100% (117) | hypothetical protein Mvan_1637 |
| M. vanbaalenii PYR-1 | Mvan_2314 | infB | 1e-05 | 59.46% (37) | translation initiation factor IF-2 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1485c | PE_PGRS27 | 1e-05 | 60.61% (33) | PE-PGRS family protein |
| M. gilvum PYR-GCK | Mflv_4805 | - | 2e-42 | 64.46% (121) | hypothetical protein Mflv_4805 |
| M. tuberculosis H37Rv | Rv2839c | infB | 5e-05 | 34.12% (85) | translation initiation factor IF-2 |
| M. leprae Br4923 | MLBr_01556 | infB | 3e-05 | 37.66% (77) | translation initiation factor IF-2 |
| M. abscessus ATCC 19977 | MAB_1609c | - | 2e-21 | 49.49% (99) | hypothetical protein MAB_1609c |
| M. marinum M | MMAR_1894 | infB | 8e-07 | 62.16% (37) | translation initiation factor IF-2 InfB |
| M. avium 104 | MAV_3693 | infB | 9e-09 | 60.53% (38) | translation initiation factor IF-2 |
| M. smegmatis MC2 155 | MSMEG_5499 | - | 5e-17 | 48.42% (95) | hypothetical protein MSMEG_5499 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_2122 | infB | 6e-07 | 62.16% (37) | translation initiation factor IF-2 |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_1637|M.vanbaalenii_PYR-1 ------------------MRLVYLIAVPVV-VSTGIAIAPPAVADCTSAG
Mflv_4805|M.gilvum_PYR-GCK ------------------MHMIAVVMTPVVSLATVVAVAAPAAADCTTAG
MMAR_1894|M.marinum_M ----MAGKARVHELAKELGVTSKEVLARLSEQGEFVKSASSTVEAPVARR
MUL_2122|M.ulcerans_Agy99 ----MAGKARVHELAKELGVTSKEVLARLSEQGEFVKSASSTVEAPVARR
Rv2839c|M.tuberculosis_H37Rv ---MAAGKARVHELAKELGVTSKEVLARLSEQGEFVKSASSTVEAPVARR
MLBr_01556|M.leprae_Br4923 ----MAGKARVHELAKELGVTSKEVLARLNEQGEFVKSASSTVEAPVARR
MAV_3693|M.avium_104 ----MAGKARVHELAKELGVTSKEVLARLNEQGEFVKSASSTVEAPVARR
MAB_1609c|M.abscessus_ATCC_199 -------------MRTMRCYLSAGVVIPLSALGVIVA--PTAAADCVNAN
MSMEG_5499|M.smegmatis_MC2_155 -------------MRSTQRYLITGFALALAPFGTVVVDPPPARADCTSSG
Mb1485c|M.bovis_AF2122/97 MSLVIVAPETVAAAALDVARIGSSIGVANSAAAGSTTSVLAAGADEVSAA
. . .: .
Mvan_1637|M.vanbaalenii_PYR-1 GTTICSQGE-----------------------------------------
Mflv_4805|M.gilvum_PYR-GCK ATTICSQGD-----------------------------------------
MMAR_1894|M.marinum_M LRESFGGGKSAPAKGSDTGAAKGVAKAPQKVPGVSPAAKAPDRSLDAALD
MUL_2122|M.ulcerans_Agy99 LRESFGGGKSAPAKGSDTGAAKGVAKAPQKVSGASPAAKAPDRSLDAALD
Rv2839c|M.tuberculosis_H37Rv LRESFGGSKPAPAKG--------TAKSPGKGP-----DKSLDKALDAAID
MLBr_01556|M.leprae_Br4923 LRESFGGIKPAADKG----AEQVATKAQAKRLG-----ESLDQTLDRALD
MAV_3693|M.avium_104 LRESFGGGK--AAEGAAKAPAKAAAKGDAKTAAKG-DVKAPDKALDAALD
MAB_1609c|M.abscessus_ATCC_199 GTTLCAQG------------------------------------------
MSMEG_5499|M.smegmatis_MC2_155 YATVCAQG------------------------------------------
Mb1485c|M.bovis_AF2122/97 IATLFGSHAREYQAISTQVAAFHDRFAQTLSAAVGSYVSAEATNAAPLAT
.
Mvan_1637|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_4805|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_1894|M.marinum_M KAVG-NGAPAPVPAPAPAPTPAPAPAPAQPADSGVAPPAATPAAPAASA-
MUL_2122|M.ulcerans_Agy99 KAVG-NGAPAPVPAPAPAPT--PAPAPAQPADSGVAPPAATPAAPAASA-
Rv2839c|M.tuberculosis_H37Rv MAAG-NGK--------------ATAAPAKAADSG----GAAIVSPTTPA-
MLBr_01556|M.leprae_Br4923 KAVAGNGATT----AAPVQVDHSAAVVPIVAGEGPSTAHREELAPPAGQ-
MAV_3693|M.avium_104 NAIKAGGNGE------------AAAPPAQPGGTATTPAAQATPEAPARPG
MAB_1609c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_5499|M.smegmatis_MC2_155 --------------------------------------------------
Mb1485c|M.bovis_AF2122/97 LEHNVLNALNAPTQALLGRPLIGDGAAGAPGTGQAGGAGGILWGNGGAGG
Mvan_1637|M.vanbaalenii_PYR-1 -----------------------VRGSTGGGGP---SGPYVPYPCDLNYY
Mflv_4805|M.gilvum_PYR-GCK -----------------------VRGTNSGTGPVGSSGPYVPYPCDWNYY
MMAR_1894|M.marinum_M -APAPPKAPLPGQRPAPTPGKPAAPQAPHPGMAPGARPGPVPKPGVRTPR
MUL_2122|M.ulcerans_Agy99 -APAPPKAPLPGQRPAPTPGKPAAPQAPHPGMAPGARPGPVPKQGVRTPR
Rv2839c|M.tuberculosis_H37Rv -APEPPTAVPP------------SPQAPHPGMAPGARPGPVPKPGIRTPR
MLBr_01556|M.leprae_Br4923 -PSEQPGVPLPGQQ-----GTPAAPHPGHPGMPTGPHPGPAPKPGGRPPR
MAV_3693|M.avium_104 PAAARPSAPAPGQP-----KPPAPGQPPRPGATPGPRPGPAPKPAARTPR
MAB_1609c|M.abscessus_ATCC_199 ------------------------DVRG-GGSTTSSTP-YVPYPCESDWS
MSMEG_5499|M.smegmatis_MC2_155 ------------------------EVRGTDGVPRAATP-VVPYPCEYDWY
Mb1485c|M.bovis_AF2122/97 SGAPGQVGGAGGAAGLFGTGGAGGAGGAGAAGGAGGSGGWLLGNGGVGGA
Mvan_1637|M.vanbaalenii_PYR-1 ECNEWWDFDADIDIDP----------------------------------
Mflv_4805|M.gilvum_PYR-GCK NCNEWWDVDFDVDLDP----------------------------------
MMAR_1894|M.marinum_M VGNNPFSSAQPVDRPIPRPQAPRPGAPRPGAPRPGGASPGNMPPRPGGAS
MUL_2122|M.ulcerans_Agy99 VGNNPFSSAQPVDRPIPRPQAPRPGAPRPGAPRPGGASSGNMPPRPGGAS
Rv2839c|M.tuberculosis_H37Rv VGNNPFSSAQPADRPIPRPPAPRPGTARPGVPRPG-ASPGSMPPRPGGAV
MLBr_01556|M.leprae_Br4923 VGNNPFSSAQSVARPIPRPPAPRP-----------SASPSSMSPRPGGAV
MAV_3693|M.avium_104 VGNNPFSSAQPVDRPIPRP-VPRPGAPRPGAPRPG-ASPGNMPPRPGGVG
MAB_1609c|M.abscessus_ATCC_199 CDNWEMDIILGSD-----------------------------HESP----
MSMEG_5499|M.smegmatis_MC2_155 CG-TDVDLDIGWD-----------------------------PGRP----
Mb1485c|M.bovis_AF2122/97 GGQSLLGGATGGAGGNAGLFGVGGTGGPGGPGGPGGPGGPGGPGGVGGTG
.
Mvan_1637|M.vanbaalenii_PYR-1 -----GPPGIGAPGRPDIGGPGRPGG------------------------
Mflv_4805|M.gilvum_PYR-GCK -----GRPGIGGPGGPGGPGIGAPGGP-----------------------
MMAR_1894|M.marinum_M GG---PRPPRTGAPRPGGGRPGGPGGGRSDGGGGNYRGGGGGVGAAPGGG
MUL_2122|M.ulcerans_Agy99 GG---PRPPRTGAPRPGGGRPGGPGGGRSDGGGGNYRGGGGGVGAAPGGG
Rv2839c|M.tuberculosis_H37Rv GG---ARPPRPGAPRPGG-RPGAPGAGRSDAGGGNYRGGG--VGAAPGTG
MLBr_01556|M.leprae_Br4923 GGG-GPRPPRTGVPRPGGGRPGAPVGGRSDAGGGNYRGGG--VGALPGGG
MAV_3693|M.avium_104 GP---GRPARPGAPRPGGGRPGGPGG--RDGGGGNYRGGG--VGAPPGGG
MAB_1609c|M.abscessus_ATCC_199 -----PPPTVNPSPPPDFGRPGRPGG------------------------
MSMEG_5499|M.smegmatis_MC2_155 -----DRP----------NRPNRPGG------------------------
Mb1485c|M.bovis_AF2122/97 GAGGLGGTLYGAGGHGGAGGPGPIGGVGGHGGVGGAAGLLGVGGHGGAGG
. . .
Mvan_1637|M.vanbaalenii_PYR-1 ----------------------------------------------GRPG
Mflv_4805|M.gilvum_PYR-GCK ----------------------------------------------GRPG
MMAR_1894|M.marinum_M ------------------------------FRGRPGGG--GGGGRPGQRG
MUL_2122|M.ulcerans_Agy99 ------------------------------FRGRPGGG--GGGGRPGQRG
Rv2839c|M.tuberculosis_H37Rv ------------------------------FRGRPGGG--GGG-RPGQRG
MLBr_01556|M.leprae_Br4923 S---------------------------GGFRGRPGGGGHGGGGRPGQRG
MAV_3693|M.avium_104 ----------------------------GGFRGRPGGG---GGGRPGQRG
MAB_1609c|M.abscessus_ATCC_199 ------------------------------------------------GG
MSMEG_5499|M.smegmatis_MC2_155 ------------------------------------------------GG
Mb1485c|M.bovis_AF2122/97 HGAAGIAGAAGKGGIFPDGSNGGAGGDAGDGGTGGRGGWLAGAGGAGGDG
*
Mvan_1637|M.vanbaalenii_PYR-1 G---GGGGIGGGRGR-----------------------------------
Mflv_4805|M.gilvum_PYR-GCK GGFGGGGGRGGGGRR-----------------------------------
MMAR_1894|M.marinum_M GAAGAFGRPGGAPRRGRKSKRAKRAEYENMQAPVVGGVRLPHGNGETIRL
MUL_2122|M.ulcerans_Agy99 GAAGAFGRPGGAPRRGRKSKRAKRAEYENMQAPVVGGVRLPHGNGETIRL
Rv2839c|M.tuberculosis_H37Rv GAAGAFGRPGGAPRRGRKSKRQKRQEYDSMQAPVVGGVRLPHGNGETIRL
MLBr_01556|M.leprae_Br4923 GAAGAFGRPGGAPRRGRKSKRQKRQEYDSMQAPVVGGVRLPHGNGETIRL
MAV_3693|M.avium_104 GAAGAFGRPGGAPRRGRKSKRQKRQEYDSMQAPVVGGVRLPHGNGETIRL
MAB_1609c|M.abscessus_ATCC_199 GGGGSIVRPR----------------------------------------
MSMEG_5499|M.smegmatis_MC2_155 G-----IGPR----------------------------------------
Mb1485c|M.bovis_AF2122/97 GIGGTGGAGGAGFSRALIVAGDNGGDGGNGGMGGAGGAGGPGGAGGLISL
*
Mvan_1637|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_4805|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_1894|M.marinum_M ARGASLSDFADKINAN---------------PAALVQALFNLGEMVTATQ
MUL_2122|M.ulcerans_Agy99 ARGASLSDFADKINAN---------------PAALVQALFNLGEMVTATQ
Rv2839c|M.tuberculosis_H37Rv ARGASLSDFADKIDAN---------------PAALVQALFNLGEMVTATQ
MLBr_01556|M.leprae_Br4923 ARGASLSDFAEKIDAN---------------PAALVQALFNLGEMVTATQ
MAV_3693|M.avium_104 ARGASLSDFAEKIDAN---------------PASLVQALFNLGEMVTATQ
MAB_1609c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_5499|M.smegmatis_MC2_155 --------------------------------------------------
Mb1485c|M.bovis_AF2122/97 LGGQGAGGDGGDGGAGGVGGDRGAGANGNQAVNAGAGGAGGHGGDPGAGG
Mvan_1637|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_4805|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_1894|M.marinum_M SVGDETLELLGSEMNYNVQVVSPEDEDRELLESFDLTYGEDSGDESELQT
MUL_2122|M.ulcerans_Agy99 SVGDETLELLGSEMNYNVQVVSPEDEDRELLESFDLTYGEDSGDESELQT
Rv2839c|M.tuberculosis_H37Rv SVGDETLELLGSEMNYNVQVVSPEDEDRELLESFDLSYGEDEGGEEDLQV
MLBr_01556|M.leprae_Br4923 SVGDETLELLGSEMNYNVQVVSPEDEDRELLEPFDLTYGEDQGDEDELQV
MAV_3693|M.avium_104 SVGDETLELLGSEMNYNVQVVSPEDEDRELLESFDLTYGEDEGTEEDLQT
MAB_1609c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_5499|M.smegmatis_MC2_155 --------------------------------------------------
Mb1485c|M.bovis_AF2122/97 AGGTGGAGSTIGAHGAAGASPTSGGNGGAGGNGAHFSSGGKAGGNGGAGG
Mvan_1637|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_4805|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_1894|M.marinum_M RPPVVTVMGHVDHGKTRLLDTIRKANVREGEAGGITQHIGAYQVSVDHDG
MUL_2122|M.ulcerans_Agy99 RPPVVTVMGHVDHGKTRLLDTIRKANVREGEAGGITQHIGAYQVSVDHDG
Rv2839c|M.tuberculosis_H37Rv RPPVVTVMGHVDHGKTRLLDTIRKANVREAEAGGITQHIGAYQVAVDLDG
MLBr_01556|M.leprae_Br4923 RPPVVTVMGHVDHGKTRLLDTIRKANVREAEAGGITQHIGAYQVGVDLDG
MAV_3693|M.avium_104 RPPVVTVMGHVDHGKTRLLDTIRKANVREAEQ-----------VTVEHDG
MAB_1609c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_5499|M.smegmatis_MC2_155 --------------------------------------------------
Mb1485c|M.bovis_AF2122/97 AGGLVGNGGAGGAGGNGAPGAPPSGGDPNGGGGGAGGAGGKGGDGGAQAG
Mvan_1637|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_4805|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_1894|M.marinum_M TERLITFIDTPGHEAFTAMRARGAKATDIAILVVAADDGVMPQTVEAINH
MUL_2122|M.ulcerans_Agy99 TERLITFIDTPGHEAFTAMRARGAKATDIAILVVAADDGVMPQTVEAINH
Rv2839c|M.tuberculosis_H37Rv SQRLITFIDTPGHEAFTAMRARGAKATDIAILVVAADDGVMPQTVEAINH
MLBr_01556|M.leprae_Br4923 SERLITFIDTPGHEAFTAMRARGAKATDIAILVVAADDGVMPQTVEAINH
MAV_3693|M.avium_104 VERPITFIDTPGHEAFTAMRARGAKATDIAILVVAADDGVMPQTVEAINH
MAB_1609c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_5499|M.smegmatis_MC2_155 --------------------------------------------------
Mb1485c|M.bovis_AF2122/97 DGGAGGAGGKGGNGGNGATGATGLNGLGAGADGTDGGKGGNGGAGGGGGA
Mvan_1637|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_4805|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_1894|M.marinum_M AQAADVPIVVAVNKIDKEGADPAKIRGQLTEYGLVAEDFGGDTMFVDISA
MUL_2122|M.ulcerans_Agy99 AQAADVPIVVAVNKIDKEGADPAKIRGQLTEYGLVAEDFGGDTMFVDISA
Rv2839c|M.tuberculosis_H37Rv AQAADVPIVVAVNKIDKEGADPAKIRGQLTEYGLVPEEFGGDTMFVDISA
MLBr_01556|M.leprae_Br4923 AQAADVPIVVAVNKIDKEGADPAKIRGQLTEYGLVAEDFGGDTMFIDISA
MAV_3693|M.avium_104 AQAADVPIVVAVNKIDVEGADPQKIRGQLTEYGLVPEEFGGDTMFVDISA
MAB_1609c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_5499|M.smegmatis_MC2_155 --------------------------------------------------
Mb1485c|M.bovis_AF2122/97 GGQGGKALAATHQDGSMGAGGAGGNGGAGGMGGDGGNGAKGTFDNGGDGV
Mvan_1637|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_4805|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_1894|M.marinum_M KQGTNIEALEEAVLLTADAALDLRANPDMEAQGVAIEAHLDRGRGPVATV
MUL_2122|M.ulcerans_Agy99 KQGTNIEALEEAVLLTADAALDLRANPDMEAQGVAIEAHLDRGRGPVATV
Rv2839c|M.tuberculosis_H37Rv KQGTNIEALEEAVLLTADAALDLRANPDMEAQGVAIEAHLDRGRGPVATV
MLBr_01556|M.leprae_Br4923 KVGTNIEALLEAVLLTADAALDLRANSGMEAQGVAIEAHLDRGRGPVATV
MAV_3693|M.avium_104 KQGTNIDQLLEAVLLTADAALDLRANPDMEAQGVAIEAHLDRGRGPVATV
MAB_1609c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_5499|M.smegmatis_MC2_155 --------------------------------------------------
Mb1485c|M.bovis_AF2122/97 GGNGGNGGSRGIGGAGGIGGAGSTAGADGARGATPTSGGNGGTGGNGANA
Mvan_1637|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_4805|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_1894|M.marinum_M LVQRGTLRVGDSVVAGDAYGRVRRMVDEHGEDVEEALPSRPVQVIGFTSV
MUL_2122|M.ulcerans_Agy99 LVQRGTLRVGDSVVAGDAYGRVRRMVDEHGEDVEEALPSRPVQVIGFTSV
Rv2839c|M.tuberculosis_H37Rv LVQRGTLRVGDSVVAGDAYGRVRRMVDEHGEDVEVALPSRPVQVIGFTSV
MLBr_01556|M.leprae_Br4923 LVQRGTLRIGDSVVAGDAYGRVRRMVDEHGVDIEAALPSSPVQVIGFTSV
MAV_3693|M.avium_104 LIQRGTLRVGDSIVAGDAYGRVRRMVDEHGDDVEEALPSRPVQVIGFTSV
MAB_1609c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_5499|M.smegmatis_MC2_155 --------------------------------------------------
Mb1485c|M.bovis_AF2122/97 TVAGGAGGAGGKGGNGGLVGNGGAGGKGGDGMAGVAGSSPTTAGESGTSG
Mvan_1637|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_4805|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_1894|M.marinum_M PGAGDNFLVVDEDRIARQIADRRSARKRNALAARSRKRISLEDLDSALKE
MUL_2122|M.ulcerans_Agy99 PGAGDNFLVVDEDRIARQIADRRSARKRNALAARSRKRISLEDLDSALKE
Rv2839c|M.tuberculosis_H37Rv PGAGDNFLVVDEDRIARQIADRRSARKRNALAARSRKRISLEDLDSALKE
MLBr_01556|M.leprae_Br4923 PGAGDNFLVVDEDRIARQIADRRSARKRNALAARSRKRISLEDLDSALKE
MAV_3693|M.avium_104 PGAGDNLLVVDEDRIARQIADKRSARKRNALAARSRKRISLEDLDSALKE
MAB_1609c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_5499|M.smegmatis_MC2_155 --------------------------------------------------
Mb1485c|M.bovis_AF2122/97 QNGGAGGAGGAGGRGGDFGGDGGTGGAGGNGADGGAGGNGANGANATTPG
Mvan_1637|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_4805|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_1894|M.marinum_M TSQLNLILKGDNAGTVEALEEALMGIEVDDEVALRVIDRGVGGITETNVN
MUL_2122|M.ulcerans_Agy99 TSQLNLILKGDNAGTVEALEEALMGIEVDDEVALRVIDRGVGGITETNVN
Rv2839c|M.tuberculosis_H37Rv TSQLNLILKGDNAGTVEALEEALMGIQVDDEVVLRVIDRGVGGITETNVN
MLBr_01556|M.leprae_Br4923 TSQLNLILKGDNAGTVEALEEALMGIQVDDEVALRVIDRGVGGITETNVN
MAV_3693|M.avium_104 TSQLNLILKGDNAGTVEALEEALMGIQIDDEVALRVIDRGVGGITETNVN
MAB_1609c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_5499|M.smegmatis_MC2_155 --------------------------------------------------
Mb1485c|M.bovis_AF2122/97 AKGGDGGHGGPGAQGGNGGQGGPGGLAGNLFGQNGIQGVGGSGGKGGAGG
Mvan_1637|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_4805|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_1894|M.marinum_M LASASDAIIIGFNVRAEGKATELANREGVEIRYYSVIYQAIDEIEKALRG
MUL_2122|M.ulcerans_Agy99 LASASDAIIIGFNVRAEGKATELANREGVEIRYYSVIYQAIDEIEKALRG
Rv2839c|M.tuberculosis_H37Rv LASASDAVIIGFNVRAEGKATELASREGVEIRYYSVIYQAIDEIEQALRG
MLBr_01556|M.leprae_Br4923 LASASDAIIIGFNVRAEGKATELASREGVEIRYYLVIYQAIDDIEKALRG
MAV_3693|M.avium_104 LASASDAVIIGFNVRAEGKATELANREGVEIRYYSVIYQAIDEIEKALRG
MAB_1609c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_5499|M.smegmatis_MC2_155 --------------------------------------------------
Mb1485c|M.bovis_AF2122/97 LAGDGGNGANGNFAFGDGNGGHGGNGGNPGAGGQGGSGGAGSTPGAKGAH
Mvan_1637|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_4805|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_1894|M.marinum_M MLKPIYEEVELGRAEIRALFRSSKVGLIAGCMISSGVVRRNAKARLLRDN
MUL_2122|M.ulcerans_Agy99 MLKPIYEEVELGRAEIRALFRSSKVGLIAGCMISSGVVRRNAKARLLRDN
Rv2839c|M.tuberculosis_H37Rv LLKPIYEENQLGRAEIRALFRSSKVGLIAGCLVTSGVMRRNAKARLLRDN
MLBr_01556|M.leprae_Br4923 MLKPIYEENQLGRAEIRALFRSSKVGIIAGCIISSGVVRRNAKVRLLRDN
MAV_3693|M.avium_104 MLKPIYEENQLGRAEIRAIFRSSKVGIIAGCMITSGVVRRNAKARLLRDN
MAB_1609c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_5499|M.smegmatis_MC2_155 --------------------------------------------------
Mb1485c|M.bovis_AF2122/97 GFTPTSGGDGGDGGNGGHSQVVGGNGGDGGNGGNGGSAGTGGNGGRGGDG
Mvan_1637|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_4805|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_1894|M.marinum_M IVVVENLSIHSLRREKDDVTEVREGFECGMTLGYSDLKEGDFIESYELVQ
MUL_2122|M.ulcerans_Agy99 IVVVENLSIRSLRREKDDVTEVREGFECGMTLGYSDLKEGDFIESYELVQ
Rv2839c|M.tuberculosis_H37Rv IVVAENLSIASLRREKDDVTEVRDGFECGLTLGYADIKEGDVIESYELVQ
MLBr_01556|M.leprae_Br4923 IVVTDNLTVTSLRREKDDVTEVREGFECGMTLGYSDIKEGDVIESYELVE
MAV_3693|M.avium_104 VVVSENLTINSLRREKDDVTEVREGFECGMTLGYSDIKEGDVIESYELVQ
MAB_1609c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_5499|M.smegmatis_MC2_155 --------------------------------------------------
Mb1485c|M.bovis_AF2122/97 AFGGMSANATNPGENGPNGNPGGNGGAGGAGGAGLNGGNGGAGGNGGLGG
Mvan_1637|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_4805|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_1894|M.marinum_M KDRS----------------------------------------------
MUL_2122|M.ulcerans_Agy99 KDRS----------------------------------------------
Rv2839c|M.tuberculosis_H37Rv KERA----------------------------------------------
MLBr_01556|M.leprae_Br4923 KQRA----------------------------------------------
MAV_3693|M.avium_104 KERT----------------------------------------------
MAB_1609c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_5499|M.smegmatis_MC2_155 --------------------------------------------------
Mb1485c|M.bovis_AF2122/97 FGGNGAAGANGVAVGAPGQPGGAGGHGGAGGNGGAGGNGGQGVVSDGAGG
Mvan_1637|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_4805|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_1894|M.marinum_M --------------------------------------------------
MUL_2122|M.ulcerans_Agy99 --------------------------------------------------
Rv2839c|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_01556|M.leprae_Br4923 --------------------------------------------------
MAV_3693|M.avium_104 --------------------------------------------------
MAB_1609c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_5499|M.smegmatis_MC2_155 --------------------------------------------------
Mb1485c|M.bovis_AF2122/97 AGGAGGDGGAPGDGANGGNGQGAGAFAGGGGGRGGDGGNAGNAGAGGPGG
Mvan_1637|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_4805|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_1894|M.marinum_M --------------------------------------------------
MUL_2122|M.ulcerans_Agy99 --------------------------------------------------
Rv2839c|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_01556|M.leprae_Br4923 --------------------------------------------------
MAV_3693|M.avium_104 --------------------------------------------------
MAB_1609c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_5499|M.smegmatis_MC2_155 --------------------------------------------------
Mb1485c|M.bovis_AF2122/97 TGSTAGKAGPAGSILHDGGNGGHGGHGAASGGNGGPGGHGGNGGNGGTGA
Mvan_1637|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_4805|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_1894|M.marinum_M --------------------------------------------------
MUL_2122|M.ulcerans_Agy99 --------------------------------------------------
Rv2839c|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_01556|M.leprae_Br4923 --------------------------------------------------
MAV_3693|M.avium_104 --------------------------------------------------
MAB_1609c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_5499|M.smegmatis_MC2_155 --------------------------------------------------
Mb1485c|M.bovis_AF2122/97 NGGNGGIGGTGGAGSTGAKGVLGTNEGDGGDGGRGGNGGRGGNGGQGLTG
Mvan_1637|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_4805|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_1894|M.marinum_M --------------------------------------------------
MUL_2122|M.ulcerans_Agy99 --------------------------------------------------
Rv2839c|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_01556|M.leprae_Br4923 --------------------------------------------------
MAV_3693|M.avium_104 --------------------------------------------------
MAB_1609c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_5499|M.smegmatis_MC2_155 --------------------------------------------------
Mb1485c|M.bovis_AF2122/97 AGGNGGTGGTPGNGGNGGNGASGDLVTSPGDGGGGGRGGDAGRGGDAGLG
Mvan_1637|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_4805|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_1894|M.marinum_M --------------------------------------------------
MUL_2122|M.ulcerans_Agy99 --------------------------------------------------
Rv2839c|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_01556|M.leprae_Br4923 --------------------------------------------------
MAV_3693|M.avium_104 --------------------------------------------------
MAB_1609c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_5499|M.smegmatis_MC2_155 --------------------------------------------------
Mb1485c|M.bovis_AF2122/97 GSSGPGGTPGDWGTGGTGGTGGTGGQGANGGLTGGRGGTGGNGGAGGTGG
Mvan_1637|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_4805|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_1894|M.marinum_M --------------------------------------------------
MUL_2122|M.ulcerans_Agy99 --------------------------------------------------
Rv2839c|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_01556|M.leprae_Br4923 --------------------------------------------------
MAV_3693|M.avium_104 --------------------------------------------------
MAB_1609c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_5499|M.smegmatis_MC2_155 --------------------------------------------------
Mb1485c|M.bovis_AF2122/97 TGHNGSQPGMGGNGGAGGFGGNGFAGVGGRGGMGGSGGTGGTGDAGPFGT
Mvan_1637|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_4805|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_1894|M.marinum_M --------------------------------------------------
MUL_2122|M.ulcerans_Agy99 --------------------------------------------------
Rv2839c|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_01556|M.leprae_Br4923 --------------------------------------------------
MAV_3693|M.avium_104 --------------------------------------------------
MAB_1609c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_5499|M.smegmatis_MC2_155 --------------------------------------------------
Mb1485c|M.bovis_AF2122/97 GTGGTGGHGGQGGGGGFSILLGLGGLGGLGSPGSIATGTAGGAGGGGGFG
Mvan_1637|M.vanbaalenii_PYR-1 --------
Mflv_4805|M.gilvum_PYR-GCK --------
MMAR_1894|M.marinum_M --------
MUL_2122|M.ulcerans_Agy99 --------
Rv2839c|M.tuberculosis_H37Rv --------
MLBr_01556|M.leprae_Br4923 --------
MAV_3693|M.avium_104 --------
MAB_1609c|M.abscessus_ATCC_199 --------
MSMEG_5499|M.smegmatis_MC2_155 --------
Mb1485c|M.bovis_AF2122/97 GLGGGEFV