For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
DDPAALTRRAADALRERTGSDAHDVAVVLGSGWAPAAAQIGDPTAAIPMAELPGFTPPSAQGHGGQVLSL RVGAHRVLVLLGRIHAYEGHDLRHVVHPVRTACAAGVHTVVLTNAAGGLRADFSVGQPVLISDHLNLTAR SPLVGAQFVDLVDAYSPRLRALAREIEPGLAEGVYAGLPGPHYETPAEIRMLRTLGADLVGMSTVHETIA ARGAGASVLGLSLVTNMAAGMTGRPLSHAEVLEAGRQSATRMGSLLSAVIARL*
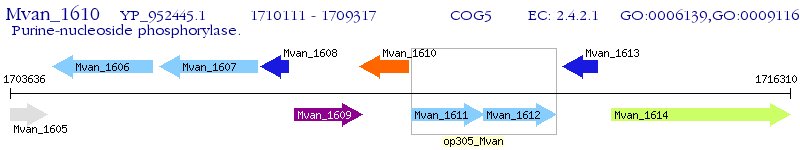
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_1610 | - | - | 100% (264) | purine nucleoside phosphorylase |
| M. vanbaalenii PYR-1 | Mvan_0881 | - | 5e-08 | 29.89% (184) | 5'-methylthioadenosine phosphorylase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3335 | deoD | 1e-113 | 77.01% (261) | purine nucleoside phosphorylase |
| M. gilvum PYR-GCK | Mflv_4828 | - | 1e-127 | 84.85% (264) | purine nucleoside phosphorylase |
| M. tuberculosis H37Rv | Rv3307 | deoD | 1e-113 | 77.01% (261) | purine nucleoside phosphorylase |
| M. leprae Br4923 | MLBr_00707 | deoD | 1e-106 | 72.80% (261) | purine nucleoside phosphorylase |
| M. abscessus ATCC 19977 | MAB_3660 | - | 1e-113 | 78.49% (265) | purine nucleoside phosphorylase |
| M. marinum M | MMAR_1219 | deoD | 1e-114 | 78.24% (262) | purine nucleoside phosphorylase DeoD |
| M. avium 104 | MAV_4284 | - | 1e-112 | 74.43% (262) | purine nucleoside phosphorylase |
| M. smegmatis MC2 155 | MSMEG_1701 | - | 1e-120 | 79.92% (264) | purine nucleoside phosphorylase |
| M. thermoresistible (build 8) | TH_2259 | deoD | 1e-119 | 78.41% (264) | PROBABLE PURINE NUCLEOSIDE PHOSPHORYLASE DEOD (INOSINE |
| M. ulcerans Agy99 | MUL_2673 | deoD | 1e-114 | 77.86% (262) | purine nucleoside phosphorylase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1219|M.marinum_M -------------------MTGPQPDPDELARRAAQVIGERTGIAEHDVA
MUL_2673|M.ulcerans_Agy99 -------------------MTGPQPDPDELARRAAQVIGERTGIAEHDVA
MLBr_00707|M.leprae_Br4923 -------------------MTYTLLDPDELARRAAQVIGERTGILKHDVA
Mb3335|M.bovis_AF2122/97 -------------------MADPRPDPDELARRAAQVIADRTGIGEHDVA
Rv3307|M.tuberculosis_H37Rv -------------------MADPRPDPDELARRAAQVIADRTGIGEHDVA
MAV_4284|M.avium_104 -------------------MAETPSNPGELARQAAAVIGERTGVAEHDVA
Mvan_1610|M.vanbaalenii_PYR-1 --------------------MDD---PAALTRRAADALRERTGSDAHDVA
Mflv_4828|M.gilvum_PYR-GCK --------------------MDD---PGVLAQQAADELRRRTGVDTHDVA
MAB_3660|M.abscessus_ATCC_1997 MTDEAPARGRYPQPARRVDDADDRLAPDDAAALAAAEIATRTGVPAHRVA
MSMEG_1701|M.smegmatis_MC2_155 --------------------MTD---PHTSSQAAATAIADRTGVVEHDVA
TH_2259|M.thermoresistible__bu --------------------VTD---PDTAATQAAAAIADRTGVPHHDVA
* : ** : *** * **
MMAR_1219|M.marinum_M VVLGSGWSAAIAALGSPTAVLAQTEVPGFTPPSAAGHTGELLSVPVG--D
MUL_2673|M.ulcerans_Agy99 VVLGSGWSAAIAALGSPTAVLAQTEVPGFTPPSAAGHAGELLSVPVG--D
MLBr_00707|M.leprae_Br4923 VVLGSGWSSAVAALGSSRAVFPQAELPGFITPNAAGHTGELLSVRIG--A
Mb3335|M.bovis_AF2122/97 VVLGSGWLPAVAALGSPTTVLPQAELPGFVPPTAAGHAGELLSVPIG--A
Rv3307|M.tuberculosis_H37Rv VVLGSGWLPAVAALGSPTTVLPQAELPGFVPPTAAGHAGELLSVPIG--A
MAV_4284|M.avium_104 IVLGSGWSPAVAALGTPTAVLPQAELPGFRPPTAVGHTGELVSMRIG--E
Mvan_1610|M.vanbaalenii_PYR-1 VVLGSGWAPAAAQIGDPTAAIPMAELPGFTPPSAQGHGGQVLSLRVG--A
Mflv_4828|M.gilvum_PYR-GCK VVLGSGWAPAATSLGEPDAAIPMADLPGFTPPSAQGHGGQVLSVRAG--P
MAB_3660|M.abscessus_ATCC_1997 VVLGSGWAPAAGELGTPATTIPMAELPAFSPPSAAGHGGSVLSVPIGGSD
MSMEG_1701|M.smegmatis_MC2_155 VVLGSGWAPAVAELGDPVGVVDMADLPGFTPPTAAGHGGQVLSVRIG--E
TH_2259|M.thermoresistible__bu VVLGSGWAPAVTELGEPVATVPMAELPGFTPPTAAGHRGQLMSTRIG--E
:****** .* :* . .. :::*.* .*.* ** *.::* *
MMAR_1219|M.marinum_M HRVLVLAGRIHAYEGHDLRHVVHPVRTACAAGARTVVLTNAAGGLRPDMQ
MUL_2673|M.ulcerans_Agy99 HRVLVLAGRIHAYEGHDLRHVVHPVRTACAAGARTVVLTNAAGGLRPDMQ
MLBr_00707|M.leprae_Br4923 HRVLVLAGRIHPYEGHDLRHVVHPVRTACAAGARIIVLTNAAGGLRADMA
Mb3335|M.bovis_AF2122/97 HRVLVLAGRIHAYEGHDLRYVVHPVRAARAAGAQIMVLTNAAGGLRADLQ
Rv3307|M.tuberculosis_H37Rv HRVLVLAGRIHAYEGHDLRYVVHPVRAARAAGAQIMVLTNAAGGLRADLQ
MAV_4284|M.avium_104 HRVLVLVGRIHAYEGHDLCHVVHPVRAACAAGVRAVVLTNAAGGLRPDLA
Mvan_1610|M.vanbaalenii_PYR-1 HRVLVLLGRIHAYEGHDLRHVVHPVRTACAAGVHTVVLTNAAGGLRADFS
Mflv_4828|M.gilvum_PYR-GCK HRVLVLLGRIHAYEGHNLRHVVHPVRTACAAGARTVVLTNAAGGVREDMT
MAB_3660|M.abscessus_ATCC_1997 ERMLILLGRIHAYEGHDLRHVVHPVRTACAAGARTIILTNAAGGLREDYA
MSMEG_1701|M.smegmatis_MC2_155 HRVLVLVGRIHAYEGHDLAHVVHPVRTAVATGVRTVVLTNAAGGLRPEYT
TH_2259|M.thermoresistible__bu HRALVLIGRIHAYEGHDLSHVVHPVRAACASGVHTVVLTNAAGGLREDFT
.* *:* ****.****:* :******:* *:*.: ::*******:* :
MMAR_1219|M.marinum_M VGQPVLISDHLNLTARSPLVGAQFVDLTDAYSARLRAVARQADPELAEGV
MUL_2673|M.ulcerans_Agy99 VGQPVLISDHLNLTARSPLVGAQFVDLTDAYSARLRAVARQADPELAEGV
MLBr_00707|M.leprae_Br4923 VGQLVLISDHLNLTTRSPLVGTHFVDLTNAYTTRLRKLASDTDPTLTEGV
Mb3335|M.bovis_AF2122/97 VGQPVLISDHLNLTARSPLVGGEFVDLTDAYSPRLRELARQSDPQLAEGV
Rv3307|M.tuberculosis_H37Rv VGQPVLISDHLNLTARSPLVGGEFVDLTDAYSPRLRELARQSDPQLAEGV
MAV_4284|M.avium_104 VGEPVLISDHLNLTGRSPLVGPQFVDLTDAYSPRLRELARQADPTLAEGV
Mvan_1610|M.vanbaalenii_PYR-1 VGQPVLISDHLNLTARSPLVGAQFVDLVDAYSPRLRALAREIEPGLAEGV
Mflv_4828|M.gilvum_PYR-GCK VGQPVLISDHLNLTARSPLVGAQFVDMVDAYSPALRALARDIDPSLTEGV
MAB_3660|M.abscessus_ATCC_1997 VGQPVLISDHLNLTARSPLVGAQFVDLVDAYSPRLRALAREIDDNLTEGV
MSMEG_1701|M.smegmatis_MC2_155 VGQPVLISDHLNLTARSPLVGPQFVDLVDAYSPRLRAVAREFDPSLAEGV
TH_2259|M.thermoresistible__bu VGQPVLISDHLNLTARSPLVGPQFVDLVDAYSPRLRRIALGIDPSLAEGV
**: ********** ****** .***:.:**:. ** :* : *:***
MMAR_1219|M.marinum_M YAGLPGPHYETPAEIRMLRVLGADLVGMSTVHETIAARAAGAEVLGVSLV
MUL_2673|M.ulcerans_Agy99 YAGLPGPHYETPAEIRMLRVLGADLVGMSTMHETIAARAAGAEVLGVSLV
MLBr_00707|M.leprae_Br4923 YAAQPGPHYETPAEIRMLRMLGADLVGMSTVHETIAARAAGAEVLGVSLV
Mb3335|M.bovis_AF2122/97 YAGLPGPHYETPAEIRMLQTLGADLVGMSTVHETIAARAAGAEVLGVSLV
Rv3307|M.tuberculosis_H37Rv YAGLPGPHYETPAEIRMLQTLGADLVGMSTVHETIAARAAGAEVLGVSLV
MAV_4284|M.avium_104 YAGLPGPHYETPAEIRMLRTLGADLVGMSTVHETIATRAAGAEVLGVSLV
Mvan_1610|M.vanbaalenii_PYR-1 YAGLPGPHYETPAEIRMLRTLGADLVGMSTVHETIAARGAGASVLGLSLV
Mflv_4828|M.gilvum_PYR-GCK YAGLPGPHYETPAEIRMLRTLGADLVGMSTVHETIAARACGAAVLGVSLV
MAB_3660|M.abscessus_ATCC_1997 YAGLPGPHYETPAEIRMLRTLGADLVGMSTVQETIAARAAGAEVLGVSLV
MSMEG_1701|M.smegmatis_MC2_155 YAGLPGPHYETPAEIRMLRTLGADLVGMSTVHETIAARAAGAEVLAVSLV
TH_2259|M.thermoresistible__bu YAGLPGPHYETPAEIRMLRTLGADLVGMSTVHETIAARAAGAQVLGLSLV
**. **************: **********::****:*..** **.:***
MMAR_1219|M.marinum_M TNLAAGITGEPLSHAEVLAAGAASAARIGSLLATVIAGLRNSDA
MUL_2673|M.ulcerans_Agy99 TNLAAGITGEPLSHAEVLAAGAASAARIGSLLATVIAGLRNSDA
MLBr_00707|M.leprae_Br4923 TNLAAGITGKPLNHAEVLAAGTASANRIGSLLADIIARF-----
Mb3335|M.bovis_AF2122/97 TNLAAGITGEPLSHAEVLAAGAASATRMGALLADVIARF-----
Rv3307|M.tuberculosis_H37Rv TNLAAGITGEPLSHAEVLAAGAASATRMGALLADVIARF-----
MAV_4284|M.avium_104 TNLAAGISGEPLSHTEVLAAGAASATRMGALLALILRQLPRF--
Mvan_1610|M.vanbaalenii_PYR-1 TNMAAGMTGRPLSHAEVLEAGRQSATRMGSLLSAVIARL-----
Mflv_4828|M.gilvum_PYR-GCK TNLAAGMTGQPLSHEEVLAAGRQSATRMGSLLSAVIAGL-----
MAB_3660|M.abscessus_ATCC_1997 TNLAAGMTGESLNHEEVLAAGRASATRMGQLLRAVIGRL-----
MSMEG_1701|M.smegmatis_MC2_155 TNLAAGMTGEPLSHAEVLQAGRESATRMGALLASVIARL-----
TH_2259|M.thermoresistible__bu TNLAAGMTGEPLSHADVLAAGRESAARMGALLATVIAAM-----
**:***::*..*.* :** ** ** *:* ** :: :