For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TPTVRTAVQEWLSHDPDPASAAELAGCSEDELAERFAQPLTFGTAGLRGPLRAGPNGMNLAVVLRATWAV ARVLTDRGLGGSQVVVGYDARHRSAEFGRAAAEVFAAQGFSVTLMFAAVPTPAVAFAVRGTGAAAGVQIT ASHNPPQDNGYKVYFPGGLQIVSPTDRDIEHAIASAPPADEIPRSPVTASGEDQVRAYLERAASVRRTTG PARIALTPMHGVGGEFALDALALAGFDDVHVVQRQFAPDPDFPTVAFPNPEEPGASDLLLALAADVDADI AIALDPDADRCAIGVPTPAGWRMLSGDETGWLLGDYILSDRQDKAVVASTVVSSRMLASIAAAHGARHVE TLTGFKWLARADEGLDARLVYAYEEAIGHCVDPDAVRDKDGISAAVLACDLVVALRGRGRTVPDALDDLA RRHGVHTTTAVTRRVDSPQDAAAMMTRLRAAPPDTIAGVAVTVADLQPRTDALVFTGGDDAASVRVVIRP SGTEPKLKSYIEVRCAGELAPGRARAAALQEAVAAAVRAWT*
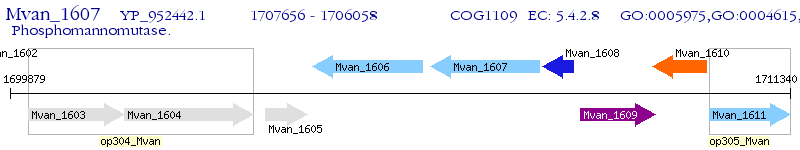
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_1607 | - | - | 100% (532) | phosphoglucomutase/phosphomannomutase alpha/beta/subunit |
| M. vanbaalenii PYR-1 | Mvan_1472 | glmM | 1e-22 | 28.34% (494) | phosphoglucosamine mutase |
| M. vanbaalenii PYR-1 | Mvan_1950 | - | 3e-19 | 26.58% (523) | phosphoglucomutase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3336 | pmmB | 0.0 | 65.29% (533) | phosphomannomutase |
| M. gilvum PYR-GCK | Mflv_4831 | - | 0.0 | 83.90% (528) | phosphoglucomutase/phosphomannomutase alpha/beta/subunit |
| M. tuberculosis H37Rv | Rv3308 | pmmB | 0.0 | 65.29% (533) | phosphomannomutase |
| M. leprae Br4923 | MLBr_00706 | pmmB | 0.0 | 65.73% (534) | putative phospho-sugar mutase |
| M. abscessus ATCC 19977 | MAB_3661 | - | 0.0 | 66.14% (505) | phosphomannomutase PmmB |
| M. marinum M | MMAR_1218 | pmmB | 0.0 | 66.54% (526) | phosphomannomutase PmmB |
| M. avium 104 | MAV_4286 | - | 0.0 | 68.50% (527) | phosphoglucomutase/phosphomannomutase |
| M. smegmatis MC2 155 | MSMEG_1695 | - | 0.0 | 68.69% (543) | phosphoglucomutase/phosphomannomutase |
| M. thermoresistible (build 8) | TH_2260 | pmmB | 0.0 | 69.07% (540) | PROBABLE PHOSPHOMANNOMUTASE PMMB (PHOSPHOMANNOSE MUTASE) |
| M. ulcerans Agy99 | MUL_2674 | pmmB | 0.0 | 66.54% (526) | phosphomannomutase PmmB |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_1607|M.vanbaalenii_PYR-1 -MTPTVRTAVQEWLSHDPDPASAAELAGCSEDELAERFAQPLTFGTAGLR
Mflv_4831|M.gilvum_PYR-GCK -----MHTAVEEWLAHDPDPESAAELAACDDDELAERFAHTLTFGTAGLR
MSMEG_1695|M.smegmatis_MC2_155 MTS--TATTAQEWIAHDPDPETAAELSACSPEELEHRFSHPLTFGTAGLR
TH_2260|M.thermoresistible__bu MTAQRTSSAVQDWIAHDPDPVTAAELRACSPEELERRFARPLRFGTAGLR
MMAR_1218|M.marinum_M -------MTPEEWIAHDPDPVTAAELAACDAQELAARFARPLAFGTAGLR
MUL_2674|M.ulcerans_Agy99 -------MTPEEWIAHDPDPVTAAELAACDAQELAARFARPLAFGTAGLR
Mb3336|M.bovis_AF2122/97 -------MTPENWIAHDPDPQTAAELAACGPDELKARFSRPLAFGTAGLR
Rv3308|M.tuberculosis_H37Rv -------MTPENWIAHDPDPQTAAELAACGPDELKARFSRPLAFGTAGLR
MLBr_00706|M.leprae_Br4923 ---MKAAVTPEEWITHDPDPQTAAELAACDPDELAARFTRALRFGTSGLR
MAV_4286|M.avium_104 -------MTPEEWIAHDPDPRTAAELAACDAAELAARFARPLRFGTAGLR
MAB_3661|M.abscessus_ATCC_1997 --MTVAAETIQEWIDHDPDPQTVAELRACSEDELAQRFSDPLVFGTAGLR
: ::*: ***** :.*** .*. ** **: .* ***:***
Mvan_1607|M.vanbaalenii_PYR-1 GPLRAGPNGMNLAVVLRATWAVARVLTDRGLGG-SQVVVGYDARHRSAEF
Mflv_4831|M.gilvum_PYR-GCK GPLRAGPNGMNLAVVIRATWAVARVLTDRSPAS-SQVVVGYDARHRSAEF
MSMEG_1695|M.smegmatis_MC2_155 GPLRGGPDGMNLAVVLRTTWAVAQVLKEHGLGE-SQVVVGRDARHKSDEF
TH_2260|M.thermoresistible__bu GPLRGGPAGMNLAVVLRATWAVAQVLKARGHAG-SGVIVGRDARHHSDEF
MMAR_1218|M.marinum_M GPVRGGPDAMNLAVVLRATWAVARVLAEQTPAGPATVIVGRDARHGSASF
MUL_2674|M.ulcerans_Agy99 GPVRGGPDAMNLAVVLRATWAVARVLAEQTPAGPATVIVGRDARHGSASF
Mb3336|M.bovis_AF2122/97 GHLRGGPDAMNLAVVLRATWAVARVLTDRGLAG-SPVIVGRDARHGSPAF
Rv3308|M.tuberculosis_H37Rv GHLRGGPDAMNLAVVLRATWAVARVLTDRGLAG-SPVIVGRDARHGSPAF
MLBr_00706|M.leprae_Br4923 GPVRGGPDAMNLAVVLRATWAVAQVLLQRAGSRPATVIVGRDSRHGSAAF
MAV_4286|M.avium_104 GPVRGGPDAMNVAVVSRASWALAQVLKRRGPAG-ARVIVGRDARHGSAVF
MAB_3661|M.abscessus_ATCC_1997 GPVRGGPNGMNLAVVLRTTYGLAKVLKDRCLGG-STVVVGRDARHGSEKF
* :*.** .**:*** *:::.:*:** : . : *:** *:** * *
Mvan_1607|M.vanbaalenii_PYR-1 GRAAAEVFAAQGFSVTLMFAAVPTPAVAFAVRGTGAAAGVQITASHNPPQ
Mflv_4831|M.gilvum_PYR-GCK GRAAAEVFAAQGFTVQLMSGPVPTPAVAFAVRHLGAAAGVQITASHNPPT
MSMEG_1695|M.smegmatis_MC2_155 ALAAAEVLAAEGFEVQLMLAAVPTPVVAFAVRHMPAVAGIQITASHNPKT
TH_2260|M.thermoresistible__bu ALAAAEVLAAEGFSVTLMFTAVPTPVLAFAVRHNRAAAGIQITASHNPAT
MMAR_1218|M.marinum_M ATATAEVLAAEGFQVVLLPEPLPTPIVAFAVRHSGAVAGIQITASHNPAT
MUL_2674|M.ulcerans_Agy99 ATATAEILAAEGFQVVLLPEPLPTPIVAFAVRHSGAVAGIQITASHNPVT
Mb3336|M.bovis_AF2122/97 AAAAAEVLAAAGFSVLLLPDPAPTPVVAFAVRHTGAAAGIQITASHNPAT
Rv3308|M.tuberculosis_H37Rv AAAAAEVLAAAGFSVLLLPDPAPTPVVAFAVRHTGAAAGIQITASHNPAT
MLBr_00706|M.leprae_Br4923 VAATAEVLAAEGFSVLLLPNPAPTPVVAFAVRNTGAAAGIQITASHNPPT
MAV_4286|M.avium_104 ATVAAEVIAAQGFSVLLLPGPAPTPVVAFAVRHTGAAAGVQITASHNPPT
MAB_3661|M.abscessus_ATCC_1997 ALGAAEVLAAEGFTVVLLPRPLPTPVLAFAVRQVDAAAGIQITASHNPPA
:**::** ** * *: . *** :***** *.**:********
Mvan_1607|M.vanbaalenii_PYR-1 DNGYKVYFPGGLQIVSPTDRDIEHAIASAPPADEIPRSPVTASG---EDQ
Mflv_4831|M.gilvum_PYR-GCK DNGYKVYVTGGLQIVSPTDREIEAAIAAAPPADEIPRAPVEVPA---ADE
MSMEG_1695|M.smegmatis_MC2_155 DNGYKVFVDGGMQITSPTDHEIEEAISRAPHADEITRAPVTTGG---LAQ
TH_2260|M.thermoresistible__bu DNGYKVYFEGGLPIVSPVDREIEEAMADAPHADEISRRPVEPSG---VDV
MMAR_1218|M.marinum_M DNGYKVYLDGGIQIISPTDRKIEAAMAAAPPADQIARKAVTPAR---TDL
MUL_2674|M.ulcerans_Agy99 DNGYKVYLDGGIQIISPTDRKIEAAMAAAPPADQIARKPVTPAR---TDL
Mb3336|M.bovis_AF2122/97 DNGYKVYVDGGLQLLAPTDRQIEAAMATAPPADQIARKTVNPSENRASDL
Rv3308|M.tuberculosis_H37Rv DNGYKVYVDGGLQLLAPTDRQIEAAMATAPPADQIARKTVNPSENRASDL
MLBr_00706|M.leprae_Br4923 DNGYKVYFDGGIQIISPIDHQIENAMAAAPLADQITRKPVNPSENSASDL
MAV_4286|M.avium_104 DNGYKVYDGGGIQIVSPTDHQIEAAMADAPPADEIRRTPVPPAH---TDL
MAB_3661|M.abscessus_ATCC_1997 DNGYKVYWSGGAQIISPTDREIEAATAAAPFADQIPRTPVEPTG---ESL
******: ** : :* *:.** * : ** **:* * .*
Mvan_1607|M.vanbaalenii_PYR-1 VRAYLERAASVRRTTGPARIALTPMHGVGGEFALDALALAGFDDVHVVQR
Mflv_4831|M.gilvum_PYR-GCK LRAYLERAASVRHAAGAARIALTPMHGVGGEFALDALALAGFADVHVVEE
MSMEG_1695|M.smegmatis_MC2_155 IKAYLERAARVRRRSDTVRVAFTPMHGVGGEFTLDAMALAGLDDVHVVES
TH_2260|M.thermoresistible__bu IQRYVERAAHVRRTHDPVRVALTPLHGVGGEFVMDALVRAGCTDVHVVES
MMAR_1218|M.marinum_M VEHYIARAAGIRRCAGSVRVALTPMHGVGGTVAVETLRRAGFTDVHTVAT
MUL_2674|M.ulcerans_Agy99 VEHYIARAAGIRRCAGSVRVALTPMHGVGGTVAVATLRRAGFTDVHTVAT
Mb3336|M.bovis_AF2122/97 IDRYIQRAAGVRRCAGSVRVALTPLHGVGGAMAVETLRRAGFTEVHTVAT
Rv3308|M.tuberculosis_H37Rv IDRYIQRAAGVRRCAGSVRVALTPLHGVGGAMAVETLRRAGFTEVHTVAT
MLBr_00706|M.leprae_Br4923 VDHYIQRAAAVRRSNGSVRVALTPMHGVGGAVAVETLRRTGFDDVHTVAA
MAV_4286|M.avium_104 VQRYIERAAGVRRGTGSVRVALTALHGVGGTVAVDALHRAGFGRIHTVAS
MAB_3661|M.abscessus_ATCC_1997 IDAYVQRAATVRRGAGNLRVVLTALHGVGGATAVATLNAAGITDIHLVDE
: *: *** :*: . *:.:*.:***** .: :: :* :* *
Mvan_1607|M.vanbaalenii_PYR-1 QFAPDPDFPTVAFPNPEEPGASDLLLALAADVDADIAIALDPDADRCAIG
Mflv_4831|M.gilvum_PYR-GCK QFAPDPDFPTVAFPNPEEPGASDALLALAAGVDAEIAIALDPDADRCAIG
MSMEG_1695|M.smegmatis_MC2_155 QFNPDPEFPTVEFPNPEEPGATDELLALAERVGAEIAIALDPDADRCAVG
TH_2260|M.thermoresistible__bu QFAPDPDFPTVPSPNPEEPAAVEAVLKLAAEVDAEIAIALDPDADRCAVG
MMAR_1218|M.marinum_M QFEPDPDFPTVAFPNPEEPGATDALLELAAGVRADVAIALDPDADRCAVA
MUL_2674|M.ulcerans_Agy99 QFEPDPDFPTVAFPNPEEPGATDALLELAAGVRADVAIALDPDADRCAVA
Mb3336|M.bovis_AF2122/97 QFAPNPDFPTVTLPNPEEPGATDALLTLATDVDADVAIALDPDADRCAVG
Rv3308|M.tuberculosis_H37Rv QFAPNPDFPTVTLPNPEEPGATDALLTLATDVDADVAIALDPDADRCAVG
MLBr_00706|M.leprae_Br4923 QFEPDPDFPTVAFPNPEEPGATDALLALAAHVGADVAIALDPDADRCAVG
MAV_4286|M.avium_104 QFAPDPDFPTVAFPNPEEPGATDALLTLAADVDADVAIALDPDADRCAVG
MAB_3661|M.abscessus_ATCC_1997 QFQPNPDFPTVEFPNPEERGASDAVLARAAEVDAALAIALDPDADRCAIG
** *:*:**** ***** .* : :* * * * :************:.
Mvan_1607|M.vanbaalenii_PYR-1 VPTPAGWRMLSGDETGWLLGDYILSDRQ------DKAVVASTVVSSRMLA
Mflv_4831|M.gilvum_PYR-GCK VPTPTGWRMLSGDETGWLLGDYVLSHADP-----THAVVASTVVSSRMLA
MSMEG_1695|M.smegmatis_MC2_155 IPTPDGWRMLSGDETGWLLGDYILSQIEPGMV-SESTVVASTVVSSRMLA
TH_2260|M.thermoresistible__bu IPTPTGWRMLSGDETGWLLGDYILAQLDDGPV-TTSTLVASTVVSSRMLA
MMAR_1218|M.marinum_M IPTSSGWRMLSGDETGWLLGDYILSKQPP-----GDWIVASTLVSSRMLA
MUL_2674|M.ulcerans_Agy99 IPTSSGWRMLSGDETGWLLGDYILSKQPS-----GDWIVASTLVSSRMLA
Mb3336|M.bovis_AF2122/97 IPTVSGWRMLSGDETGWLLGDYILSQTDDRASPPETRVVASTVVSSRMLA
Rv3308|M.tuberculosis_H37Rv IPTVSGWRMLSGDETGWLLGDYILSQTDDRASPPETRVVASTVVSSRMLA
MLBr_00706|M.leprae_Br4923 IPTNSGWRMLSGDETGWLLGDYILSQTDK----PETAVVASTVVSSRMLP
MAV_4286|M.avium_104 IPDASGWRMLSGDETGWLLGDYLLSQPQR----ADRPVVASTVVSSRMLS
MAB_3661|M.abscessus_ATCC_1997 VPTTTGWRMLSGDETGWLLGDYILSTQPDS---SIPPVVASTVVSSRMLA
:* *****************:*: :****:******.
Mvan_1607|M.vanbaalenii_PYR-1 SIAAAHGARHVETLTGFKWLARADEG-LDARLVYAYEEAIGHCVDPDAVR
Mflv_4831|M.gilvum_PYR-GCK SIAAAHGARHVETLTGFKWLARADEG-LDATLVYAYEEAIGHCVDPGAVR
MSMEG_1695|M.smegmatis_MC2_155 SIAAAHGAHHVETLTGFKWLARAGS--PGTTLVYAYEEAIGHCVDPSSVR
TH_2260|M.thermoresistible__bu AIADAHGARHVETLTGFKWLVRADADLPGSSLVYAYEEAIGHCVDPSAVR
MMAR_1218|M.marinum_M AIAADYGVVHVQTLTGFKWLARADADRPG-TLVYAYEEAIGHCVDPTAVR
MUL_2674|M.ulcerans_Agy99 AIAADYGVVHVQTLTGFKWLARADADRPG-TLVYAYEEAIGHCVDPTAVR
Mb3336|M.bovis_AF2122/97 AIAAHHAAVHVETLTGFKWLARADANLPG-TLVYAYEEAIGHCVDPTAVR
Rv3308|M.tuberculosis_H37Rv AIAAHHAAVHVETLTGFKWLARADANLPG-TLVYAYEEAIGHCVDPTAVR
MLBr_00706|M.leprae_Br4923 AIATHYNAVHVETLTGFKWLARADANLPG-TLVYAYEEAIGHCVDPTAVR
MAV_4286|M.avium_104 AIAARHGAVHVETLTGFKWLARADAEVPGGTLVYAYEEAIGHCVDPAAVR
MAB_3661|M.abscessus_ATCC_1997 RIAASLGARHVETLTGFKWLVRAADS----DLVYAYEEAIGHCVDPVAVR
** . **:********.** *************** :**
Mvan_1607|M.vanbaalenii_PYR-1 DKDGISAAVLACDLVVALRGRGRTVPDALDDLARRHGVHTTTAVTRRVD-
Mflv_4831|M.gilvum_PYR-GCK DKDGISAAVLACDLVVSLREQGRTLLDALDDLARRHGVHTTTAVTRRVR-
MSMEG_1695|M.smegmatis_MC2_155 DKDGITAAILACDLVAALRYQGRTIVDALDGLARAHGVHVTRAVSRQVS-
TH_2260|M.thermoresistible__bu DKDGISAAVLCCDLVAALRAQGRTVLDALDDLARRFGVHTTTASSRRVE-
MMAR_1218|M.marinum_M DKDGISAAVLTCDLVASLERQGRSVPDVLDDLAHRYGVHEVGAVARRVA-
MUL_2674|M.ulcerans_Agy99 DKDGISAAVLTCDLVASLERQGRSVPDVLDDLAHRYGVHEVGAVARRVA-
Mb3336|M.bovis_AF2122/97 DKDGISAAVLVCDLVAALKGQGRSVTDALDELARCYGVHEVAALSRPVG-
Rv3308|M.tuberculosis_H37Rv DKDGISAAVLVCDLVAALKGQGRSVTDALDELARCYGVHEVAALSRPVS-
MLBr_00706|M.leprae_Br4923 DKDGISAAVLVCDLVAALHKQGRSVPDMLDQLALRHGVHDVTAISRRIGP
MAV_4286|M.avium_104 DKDGISAAVLVCDLVAALIARGGSVSGRLDELARRHGVHDVAAVSRRVG-
MAB_3661|M.abscessus_ATCC_1997 DKDGISAAVLACDLVTHLRELGTSVEEKLENLAIRHGIHVTSQVSRRMD-
*****:**:* ****. * * :: *: ** .*:* . :* :
Mvan_1607|M.vanbaalenii_PYR-1 ---SPQDAAAMMTRLRAAPPDTIAGVAVTVADLQP-----RTDALVFTGG
Mflv_4831|M.gilvum_PYR-GCK ---SPQDAAAMMSRLRAAPPEEIAGFAVTVTDLMP-----RTDALIFSGG
MSMEG_1695|M.smegmatis_MC2_155 ---DADKAMG---RLRTQPPDELAGFTVAVEDLAERRGQQRTDALIFTGE
TH_2260|M.thermoresistible__bu ---DPDQAAAMLDRLRRRPPTELTGFPVTVTDLARAEGARRTDALIFEGG
MMAR_1218|M.marinum_M ---DADQAAALMRRLRESPPDRLAGFTATVTDITD--------ALIFAGG
MUL_2674|M.ulcerans_Agy99 ---DADQAAALMRRLRESPPDRLAGFTATVTDITD--------ALIFAGG
Mb3336|M.bovis_AF2122/97 ---GAVETTDLMRRLREDPPRRLAGFPATVTDIGD--------TLILTGG
Rv3308|M.tuberculosis_H37Rv ---GAVETTDLMRRLREDPPRRLAGFPATVTDIGD--------TLILTGG
MLBr_00706|M.leprae_Br4923 KQTGVDEAVDLIQRLRAAPPSQLAGFTATTTDITD--------ALIFTGG
MAV_4286|M.avium_104 ----TDGAAELMRRLRTSPPETLAGFGVALTDITD--------ALIFTGG
MAB_3661|M.abscessus_ATCC_1997 ---SLEDIEAMMTRLRQNIPAALSGFPITAEDLLERRGQQRTDAVVLSGE
*** * ::*. : *: :::: *
Mvan_1607|M.vanbaalenii_PYR-1 DDAASVRVVIRPSGTEPKLKSYIEVR----CAGELAPGRARAAALQEAVA
Mflv_4831|M.gilvum_PYR-GCK DSATSVRVVMRPSGTEPKLKSYIEIR----CTGEVATGRSHAADVQDAVA
MSMEG_1695|M.smegmatis_MC2_155 DGGTAVRIVVRPSGTEPKLKSYIEVR--CAPTDDLAAARERANRISDELC
TH_2260|M.thermoresistible__bu TADTAVRVVVRPSGTEPKVKFYFEVRRSCHDDEDLDIARARARELQDELV
MMAR_1218|M.marinum_M DDRTSVRVVVRPSGTEPKLKCYIEVR--CEVAGDLAATRQRAQELRRELA
MUL_2674|M.ulcerans_Agy99 DDRTSVRVVVRPSGTEPKLKCYIEVR--CEVAGDLAATRQRAQELRRELA
Mb3336|M.bovis_AF2122/97 DDNMLVRVAVRPSGTEPKLKCYLEIR--CAVTGDLPAARQLVRARIDELS
Rv3308|M.tuberculosis_H37Rv DDNMLVRVAVRPSGTEPKLKCYLEIR--CAVTGDLPAARQLVRARIDELS
MLBr_00706|M.leprae_Br4923 DDDTWVRVVVRLSGTEPKLKCYLEVR--CSVAGNLPSTRQRARVLRDELV
MAV_4286|M.avium_104 DDDTSVRLVVRPSGTEPKVKCYLEIR--CAPSDDLASSRRRARALRELLV
MAB_3661|M.abscessus_ATCC_1997 IGGSSIRLVLRPSGTEPKVKCYIEVR--EPVITDPTVARIWAGVVVSELE
:*:.:* ******:* *:*:* : * . :
Mvan_1607|M.vanbaalenii_PYR-1 AAVRAWT
Mflv_4831|M.gilvum_PYR-GCK AAIATWR
MSMEG_1695|M.smegmatis_MC2_155 RAAKGW-
TH_2260|M.thermoresistible__bu ATVDQW-
MMAR_1218|M.marinum_M STVSGW-
MUL_2674|M.ulcerans_Agy99 STVSGW-
Mb3336|M.bovis_AF2122/97 ASVRRWW
Rv3308|M.tuberculosis_H37Rv ASVRRWW
MLBr_00706|M.leprae_Br4923 TLVQQW-
MAV_4286|M.avium_104 ATVRSW-
MAB_3661|M.abscessus_ATCC_1997 AYARNI-