For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RTGIFLSYAGGFLEAVDEVVECEKLGVDIALVAEAYSYDAISQLGFLAARTSRIELGTGVVPIYTRTPAL MAMTAAGLDYVSDGRFRLGLGTSGPQVVEGFHGVPFDAPLGRTREVVEICRQVWRRERLSYDGKYYQLPL PSDKGTGLGKPLKLINHPVRERIPITIAALGPKNVELTAEIAEGWQPVFFYPEKADDVWGDALRAGASRR DPALGPLDTMVSASLAIGDDVDDRLAWAKPQLALYIGGMGARGKNFYHNLATRYGFGEVADHIQDLYLAG KKAEAIDAVPDELVRQVSLVGPAGFVKERLAAYAEAGVTTMLVHPLSGDRRETVKFVEELQNLLP*
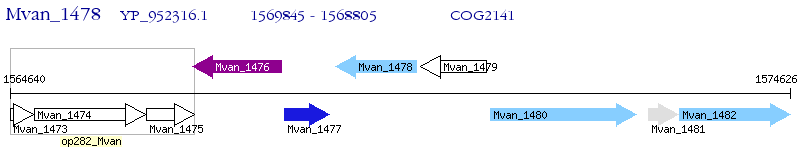
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_1478 | - | - | 100% (346) | luciferase family protein |
| M. vanbaalenii PYR-1 | Mvan_5219 | - | 2e-68 | 42.55% (329) | luciferase family protein |
| M. vanbaalenii PYR-1 | Mvan_0319 | - | 8e-19 | 23.28% (335) | luciferase family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3550c | - | 5e-66 | 42.90% (331) | coenzyme F420-dependent oxidoreductase |
| M. gilvum PYR-GCK | Mflv_4941 | - | 0.0 | 92.75% (345) | luciferase family protein |
| M. tuberculosis H37Rv | Rv3520c | - | 5e-66 | 42.90% (331) | coenzyme F420-dependent oxidoreductase |
| M. leprae Br4923 | MLBr_00348 | - | 6e-69 | 44.11% (331) | putative coenzyme F420-dependent oxidoreductase |
| M. abscessus ATCC 19977 | MAB_3748 | - | 1e-141 | 69.57% (345) | putative luciferase-like oxidoreductase |
| M. marinum M | MMAR_1111 | - | 1e-152 | 74.42% (344) | coenzyme F420-dependent oxidoreductase |
| M. avium 104 | MAV_4381 | - | 1e-159 | 76.01% (346) | oxidoreductase |
| M. smegmatis MC2 155 | MSMEG_1566 | - | 1e-173 | 86.51% (341) | oxidoreductase |
| M. thermoresistible (build 8) | TH_1703 | - | 1e-153 | 75.36% (345) | oxidoreductase |
| M. ulcerans Agy99 | MUL_4943 | - | 1e-132 | 64.35% (345) | coenzyme F420-dependent oxidoreductase |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_1478|M.vanbaalenii_PYR-1 -------MRTGIFLSYAGG--FLEAVDEVVECEKLGVDIALVAEAYSYDA
Mflv_4941|M.gilvum_PYR-GCK -------MRTGIFLSYAGG--FLEAVDEVVECEKLGVDIALVAEAYSYDA
MSMEG_1566|M.smegmatis_MC2_155 ------MMRTGIFLSYAGG--FKEAADQVVELEKVGVDIALVAEAYSFDA
TH_1703|M.thermoresistible__bu -------MRIGMQLAYSGG--FKETAGLVVELEKVGLDLAVVPEAYSFDA
MMAR_1111|M.marinum_M -------MRIGIALDYSGG--FHEGLDRVVELEKSGIDVAVVAEAYAFDA
MAV_4381|M.avium_104 -------MRIGIALNYSGG--FHEAVDRVVELEKAGIEVAVVAEAYSFDA
MAB_3748|M.abscessus_ATCC_1997 -------MRIGTTLSYAGG--FTEVVDELAELEKIGLDIAFVAEAYSYDA
MUL_4943|M.ulcerans_Agy99 -------MRIGLAINYAGG--FKEVAAEVADLERAGLDIVFVPEAYSFDA
Mb3550c|M.bovis_AF2122/97 -MEAG--MKLGLQLGYWGAQPPQNHAELVAAAEDAGFDTVFTAEAWGSDA
Rv3520c|M.tuberculosis_H37Rv -MEAG--MKLGLQLGYWGAQPPQNHAELVAAAEDAGFDTVFTAEAWGSDA
MLBr_00348|M.leprae_Br4923 MVERANAMKLGLQLGYWAAQPPGNAAELVAAAEGAGFDAVFTGEAWGSDA
*: * : * .. : :. * *.: ... **:. **
Mvan_1478|M.vanbaalenii_PYR-1 ISQLGFLAARTSRIELGTGVVPIYTRTPALMAMTAAGLDYVSDGRFRLGL
Mflv_4941|M.gilvum_PYR-GCK ISQLGFLAARTSRIELGTGVVPIYTRTPALMAMTAAGVDYVSDGRFRLGL
MSMEG_1566|M.smegmatis_MC2_155 ISQLGYLAAKTSTIELGTGVVPIYTRTPSLLAMTAAGVDYVSDGRFRLGI
TH_1703|M.thermoresistible__bu ISQLGYLAARTSTIQLATGVVPIYTRTPALLAMTAAGLDYVSDGRFTLGL
MMAR_1111|M.marinum_M VSQLGYLAAKTTRVQLASGVLPIYIRTPSLLAMTAAGLDYVSGGRFNLGI
MAV_4381|M.avium_104 ISQLGYLAAKTSTVELASGVLPLYIRTPSLLAMTAAGLDYVSDGRFRLGI
MAB_3748|M.abscessus_ATCC_1997 ASQLGYLAAKTSTVKLASGIFQLYTRTPTLLAMTAAGLDYVSGGRFVLGI
MUL_4943|M.ulcerans_Agy99 VSALGYLAACTDRVQLASGILQLYTRTPTLTAMTAAGLDYVSDGRFTLGL
Mb3550c|M.bovis_AF2122/97 YTPLAWWGSSTQRVRLGTSVIQLSARTPTACAMAALTLDHLSGGRHILGL
Rv3520c|M.tuberculosis_H37Rv YTPLAWWGSSTQRVRLGTSVIQLSARTPTACAMAALTLDHLSGGRHILGL
MLBr_00348|M.leprae_Br4923 YTPLAWWGSSTQRVRLGTSVVQLSARTPTACAMAALTLDHLSGGRHILGL
: *.: .: * :.*.:.:. : ***: **:* :*::*.**. **:
Mvan_1478|M.vanbaalenii_PYR-1 GTSGPQVVEGFHGVPFDAPLGRTREVVEICRQVWRRER-LSYDGKYYQLP
Mflv_4941|M.gilvum_PYR-GCK GTSGPQVIEGFHGLPFDAPLGRTREVVEICRKVWRRER-VSYDGKYYQLP
MSMEG_1566|M.smegmatis_MC2_155 GTSGPQVVEGFHGVPFDAPLGRTREVVEICRQVWRREN-VDFDGRYYQLP
TH_1703|M.thermoresistible__bu GTSGPQVVEGFHGVPFDAPLGRTREVVEICRKVWRRER-LQYNGRHYHLP
MMAR_1111|M.marinum_M GTSGPQVIEGFHGVEFDAPIGRTREVVEICRHVWRRET-LRYAGAHYQIP
MAV_4381|M.avium_104 GTSGPQVIEGFHGVPFDAPIGRTREIVEICRMVWRRER-VQYAGRHYQLP
MAB_3748|M.abscessus_ATCC_1997 GASGPQVIEGFHGVKYDAPLGRTRETVEICRKVWRRER-LEFEGKYFTVP
MUL_4943|M.ulcerans_Agy99 GASGPQVIEGFHGVPYDAPIARTREIIEICRQVWRRER-LEYQGKHYTIP
Mb3550c|M.bovis_AF2122/97 GVSGPQVVEGWYGQRFPKPLARTREYIDIVRQVWARESPVTSAGPHYRLP
Rv3520c|M.tuberculosis_H37Rv GVSGPQVVEGWYGQRFPKPLARTREYIDIVRQVWARESPVTSAGPHYRLP
MLBr_00348|M.leprae_Br4923 GVSGPQVVEGWYGQPFPKPLSRTREYIDIVRQVWAREAPVVSDGQHYPLP
*.*****:**::* : *:.**** ::* * ** ** : * :: :*
Mvan_1478|M.vanbaalenii_PYR-1 LPSDKGTGLGKPLKLINHPVRERIPITIAALGPKNVELTAEIAEGWQPVF
Mflv_4941|M.gilvum_PYR-GCK LPAERGTGLGKPLKLINHPVRENIPITIAALGPKNVELTAEIAEGWQPVF
MSMEG_1566|M.smegmatis_MC2_155 LPAERGTGLGKSLHLINHPVRERIPITIAALGPKNVELTAEIAEGWQPVF
TH_1703|M.thermoresistible__bu LPPDRGTGLGKPLKLINHPVREHIPIFIAALGPKNVQLTAEIADGWQPVF
MMAR_1111|M.marinum_M LPANRGTGLGKALKLINHPLRERIPISIAALGPRNVELTAEIAEGWQPVF
MAV_4381|M.avium_104 LPPDRGTGLGKPLHLINHPVRERIPISIAALGPKNVELTAEIAEGWQPVF
MAB_3748|M.abscessus_ATCC_1997 LPPEQGTGLGKALKLINHPVRDRIPVLIAALGPKNVALTAEIAEGWQPIF
MUL_4943|M.ulcerans_Agy99 LPAGQGTGLGKPLKIINQPVQKRIPILLAALGPKNVELAAEAAEGWQPIF
Mb3550c|M.bovis_AF2122/97 LTGEGTTGLGKALKPITHPLRADIPIMLGAEGPKNVALAAEICDGWLPIF
Rv3520c|M.tuberculosis_H37Rv LTGEGTTGLGKALKPITHPLRADIPIMLGAEGPKNVALAAEICDGWLPIF
MLBr_00348|M.leprae_Br4923 LTGAGATGLGKALKPITHPLRADIPIMLGAEGPKNVALAAEICDGWLPIF
*. *****.*: *.:*:: **: :.* **:** *:** .:** *:*
Mvan_1478|M.vanbaalenii_PYR-1 FYPEKADDVWGDALRAGASRRDP--ALGPLDTMVSASLAIGDDVDDRLAW
Mflv_4941|M.gilvum_PYR-GCK FYPEKADEVWGDALRAGASRRDP--ALGPLDVMVSATLAIGDDVDDRLSW
MSMEG_1566|M.smegmatis_MC2_155 YFPEKADDVWGDALRAGAAKRDP--DLDPLDVVVSASLAIGDDVDDRLAW
TH_1703|M.thermoresistible__bu VDPERIDSVWGDALRAGRAKRDP--TLGPLDVMGTPPLAIGDDVEDRIAW
MMAR_1111|M.marinum_M FYPEKAASAWGEALASGTAKRDP--ALGPLDVTVHASLAIGDDVEQRLAR
MAV_4381|M.avium_104 YLPDKAGSIWGEALAAGAAKRDP--ALGPLDVMVHASLAIGDDVDQRLAW
MAB_3748|M.abscessus_ATCC_1997 FLPEKANDVWGAALAEGKAKRDP--ALGDLEVYAGPALAIGDDVEALYGF
MUL_4943|M.ulcerans_Agy99 YLPEKAHDVWGQALAAGRAKRDP--ALGELDVYAGPVLAIGDNVESLREF
Mb3550c|M.bovis_AF2122/97 YSP-RMAGMYNEWLDEGFARPGARRSREDFEICATAQVVITDDRAAAFAG
Rv3520c|M.tuberculosis_H37Rv YSP-RMAGMYNEWLDEGFARPGARRSREDFEICATAQVVITDDRAAAFAG
MLBr_00348|M.leprae_Br4923 FSP-RMADMYNEWLDEGFARTGARRSREDFEICASAQIVVTEDRAAAFAG
* : :. * * :: .. :: . :.: ::
Mvan_1478|M.vanbaalenii_PYR-1 AKPQLALYIGGMGARGKNFYHNLATRYGFGEVADHIQDLYLAGKKAEAID
Mflv_4941|M.gilvum_PYR-GCK GKPQLALYIGGMGARGKNFYHNLATRYGFGEVADHIQDLYLAGKKAEAID
MSMEG_1566|M.smegmatis_MC2_155 AKPQLALYIGGMGARGKNFYHNLATRYGYGEVADQIQDLYLAGKKAEAIA
TH_1703|M.thermoresistible__bu VKPMLALYLGGMGARDKNFYHNLATRYGFGAEADRIQELYLAGRKEEAAA
MMAR_1111|M.marinum_M VKPQLALYLGGMGAKGRNFYHNLATRYGYGDVADRIQELYLAGRKHEAIA
MAV_4381|M.avium_104 VKPQLALYIGGMGAKGRNFYHNLATRYGFGEVADRIQELYLSGRKQQAID
MAB_3748|M.abscessus_ATCC_1997 VKPSLALYIGGMGAKGKNFYHNLATAYGFGKEADTIQELYLSGKKAEATD
MUL_4943|M.ulcerans_Agy99 VKPHLALYIGGMGAKGKNFYHMLATRYGYGAEADRIQQLYLAGDKDGAAK
Mb3550c|M.bovis_AF2122/97 IKPFLALYMGGMGAEETNFHADVYRRMGYTQVVDEVTKLFRSGRKDEAAE
Rv3520c|M.tuberculosis_H37Rv IKPFLALYMGGMGAEETNFHADVYRRMGYTQVVDEVTKLFRSGRKDEAAE
MLBr_00348|M.leprae_Br4923 IKPFLALYMGGMGAEGTNFHADVYRRMGYAEVVDEVTALFRSNQKDKAAE
** ****:*****. **: : *: .* : *: :. * *
Mvan_1478|M.vanbaalenii_PYR-1 AVPDELVRQVSLVGPAGFVKERLAAYAEAGVTTMLVHPLSGDRRETVKFV
Mflv_4941|M.gilvum_PYR-GCK AVPDELVRQVSLVGPAGFVKERLAAFAEAGVTTLLVHPMGVDAAERVRFV
MSMEG_1566|M.smegmatis_MC2_155 AVPDDLVRNVSLVGPRGFVKERLAAYAESGVTTLLVHPLATDTAEAVRFC
TH_1703|M.thermoresistible__bu AVPDDLVRSVSLIGPAGLVAERVAMFAEAGVTTLLVTPLATDTAEAVRFI
MMAR_1111|M.marinum_M AVPDELARGISLIGPRGFVAERLAAFAEAGVTTMLVSPLSTDRREYVRHV
MAV_4381|M.avium_104 LVPDELVRGMSLVGPRGYVAERMAAFAESGVTTLLLSPLAADRDEALGFV
MAB_3748|M.abscessus_ATCC_1997 AVPDELVKNISLIGPKSFVAERVAAFKEAGVTTLNVAPLAADAAGRIKHV
MUL_4943|M.ulcerans_Agy99 VVPDELVRDVNLIGTPGFVRDRLAAFRESGVTTLNVAPIAETAAERVKLI
Mb3550c|M.bovis_AF2122/97 IIPDELVDDAVIVGDIDHVRKQMAVWEAAGVTMMVVT---AGSAEQVRDL
Rv3520c|M.tuberculosis_H37Rv IIPDELVDDAVIVGDIDHVRKQMAVWEAAGVTMMVVT---AGSAEQVRDL
MLBr_00348|M.leprae_Br4923 IIPNELVDSTMIVGDVDYVCKQIAAWSAAGVTMMMVS---ASSVEQVRDL
:*::*. ::* . * .::* : :*** : : :
Mvan_1478|M.vanbaalenii_PYR-1 EELQNLLP-
Mflv_4941|M.gilvum_PYR-GCK EQLQGLL--
MSMEG_1566|M.smegmatis_MC2_155 EELVALTR-
TH_1703|M.thermoresistible__bu EQLRTMVA-
MMAR_1111|M.marinum_M EQLLGLRRA
MAV_4381|M.avium_104 EQALQLRP-
MAB_3748|M.abscessus_ATCC_1997 EALRELL--
MUL_4943|M.ulcerans_Agy99 DTLRQLVD-
Mb3550c|M.bovis_AF2122/97 AALV-----
Rv3520c|M.tuberculosis_H37Rv AALV-----
MLBr_00348|M.leprae_Br4923 AGLF-----