For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RIGLAINYAGGFKEVAAEVADLERAGLDIVFVPEAYSFDAVSALGYLAACTDRVQLASGILQLYTRTPTL TAMTAAGLDYVSDGRFTLGLGASGPQVIEGFHGVPYDAPIARTREIIEICRQVWRRERLEYQGKHYTIPL PAGQGTGLGKPLKIINQPVQKRIPILLAALGPKNVELAAEAAEGWQPIFYLPEKAHDVWGQALAAGRAKR DPALGELDVYAGPVLAIGDNVESLREFVKPHLALYIGGMGAKGKNFYHMLATRYGYGAEADRIQQLYLAG DKDGAAKVVPDELVRDVNLIGTPGFVRDRLAAFRESGVTTLNVAPIAETAAERVKLIDTLRQLVD*
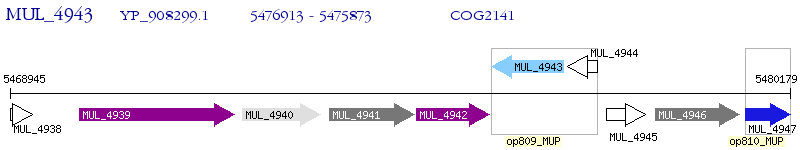
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_4943 | - | - | 100% (346) | coenzyme F420-dependent oxidoreductase |
| M. ulcerans Agy99 | MUL_4079 | - | 7e-67 | 42.12% (330) | coenzyme F420-dependent oxidoreductase |
| M. ulcerans Agy99 | MUL_4782 | - | 8e-66 | 39.71% (350) | coenzyme F420-dependent oxidoreductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3550c | - | 4e-67 | 42.65% (347) | coenzyme F420-dependent oxidoreductase |
| M. gilvum PYR-GCK | Mflv_4941 | - | 1e-132 | 64.64% (345) | luciferase family protein |
| M. tuberculosis H37Rv | Rv3520c | - | 4e-67 | 42.65% (347) | coenzyme F420-dependent oxidoreductase |
| M. leprae Br4923 | MLBr_00348 | - | 4e-67 | 41.82% (330) | putative coenzyme F420-dependent oxidoreductase |
| M. abscessus ATCC 19977 | MAB_3748 | - | 1e-145 | 71.88% (345) | putative luciferase-like oxidoreductase |
| M. marinum M | MMAR_0154 | - | 0.0 | 99.13% (346) | coenzyme F420-dependent oxidoreductase |
| M. avium 104 | MAV_0137 | - | 1e-172 | 85.22% (345) | oxidoreductase |
| M. smegmatis MC2 155 | MSMEG_6602 | - | 1e-173 | 85.80% (345) | oxidoreductase |
| M. thermoresistible (build 8) | TH_1703 | - | 1e-137 | 67.25% (345) | oxidoreductase |
| M. vanbaalenii PYR-1 | Mvan_1478 | - | 1e-132 | 64.35% (345) | luciferase family protein |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_4943|M.ulcerans_Agy99 -------MRIGLAINYAGG--FKEVAAEVADLERAGLDIVFVPEAYSFDA
MMAR_0154|M.marinum_M -------MRIGLAINYAGG--FKEVAAEVADLERAGLDIVFVPEAYSFDA
MAV_0137|M.avium_104 -----MTMRIGLSINYAGG--FKDVAAEVADLENAGLDIVFVPEAYSFDA
MSMEG_6602|M.smegmatis_MC2_155 -------MKIGLSINYAGG--FKDIAAEVADLERAGLDIVFVPEAYSFDA
MAB_3748|M.abscessus_ATCC_1997 -------MRIGTTLSYAGG--FTEVVDELAELEKIGLDIAFVAEAYSYDA
Mflv_4941|M.gilvum_PYR-GCK -------MRTGIFLSYAGG--FLEAVDEVVECEKLGVDIALVAEAYSYDA
Mvan_1478|M.vanbaalenii_PYR-1 -------MRTGIFLSYAGG--FLEAVDEVVECEKLGVDIALVAEAYSYDA
TH_1703|M.thermoresistible__bu -------MRIGMQLAYSGG--FKETAGLVVELEKVGLDLAVVPEAYSFDA
Mb3550c|M.bovis_AF2122/97 -MEAG--MKLGLQLGYWGAQPPQNHAELVAAAEDAGFDTVFTAEAWGSDA
Rv3520c|M.tuberculosis_H37Rv -MEAG--MKLGLQLGYWGAQPPQNHAELVAAAEDAGFDTVFTAEAWGSDA
MLBr_00348|M.leprae_Br4923 MVERANAMKLGLQLGYWAAQPPGNAAELVAAAEGAGFDAVFTGEAWGSDA
*: * : * .. : . :. * *.* ... **:. **
MUL_4943|M.ulcerans_Agy99 VSALGYLAACTDRVQLASGILQLYTRTPTLTAMTAAGLDYVSDGRFTLGL
MMAR_0154|M.marinum_M VSALGYLAACTDRVQLASGILQLYTRTPTLTAMTAAGLDYVSDGRFTLGL
MAV_0137|M.avium_104 VSALGYLAASTQRVELASGILQLYTRTPTLTAMTAAGLDYVSDGRFTLGL
MSMEG_6602|M.smegmatis_MC2_155 VSGLGYLAARTDRVQLASGILQLYTRTPTLTAMTAAGLDYVSDGRFILGL
MAB_3748|M.abscessus_ATCC_1997 ASQLGYLAAKTSTVKLASGIFQLYTRTPTLLAMTAAGLDYVSGGRFVLGI
Mflv_4941|M.gilvum_PYR-GCK ISQLGFLAARTSRIELGTGVVPIYTRTPALMAMTAAGVDYVSDGRFRLGL
Mvan_1478|M.vanbaalenii_PYR-1 ISQLGFLAARTSRIELGTGVVPIYTRTPALMAMTAAGLDYVSDGRFRLGL
TH_1703|M.thermoresistible__bu ISQLGYLAARTSTIQLATGVVPIYTRTPALLAMTAAGLDYVSDGRFTLGL
Mb3550c|M.bovis_AF2122/97 YTPLAWWGSSTQRVRLGTSVIQLSARTPTACAMAALTLDHLSGGRHILGL
Rv3520c|M.tuberculosis_H37Rv YTPLAWWGSSTQRVRLGTSVIQLSARTPTACAMAALTLDHLSGGRHILGL
MLBr_00348|M.leprae_Br4923 YTPLAWWGSSTQRVRLGTSVVQLSARTPTACAMAALTLDHLSGGRHILGL
: *.: .: *. :.*.:.:. : :***: **:* :*::*.**. **:
MUL_4943|M.ulcerans_Agy99 GASGPQVIEGFHGVPYDAPIARTREIIEICRQVWRRE-RLEYQGKHYTIP
MMAR_0154|M.marinum_M GASGPQVIEGFHGVPYDAPIARTREIIEICRQVWRRE-RLEYQGKHYTIP
MAV_0137|M.avium_104 GASGPQVIEGFHGVPYDAPIGRTREVIEICRQVWRRE-PVQHRGKHYTIP
MSMEG_6602|M.smegmatis_MC2_155 GASGPQVIEGFHGVRYDAPIARTREVVEICRRVWRRE-RLQYEGKHYTVP
MAB_3748|M.abscessus_ATCC_1997 GASGPQVIEGFHGVKYDAPLGRTRETVEICRKVWRRE-RLEFEGKYFTVP
Mflv_4941|M.gilvum_PYR-GCK GTSGPQVIEGFHGLPFDAPLGRTREVVEICRKVWRRE-RVSYDGKYYQLP
Mvan_1478|M.vanbaalenii_PYR-1 GTSGPQVVEGFHGVPFDAPLGRTREVVEICRQVWRRE-RLSYDGKYYQLP
TH_1703|M.thermoresistible__bu GTSGPQVVEGFHGVPFDAPLGRTREVVEICRKVWRRE-RLQYNGRHYHLP
Mb3550c|M.bovis_AF2122/97 GVSGPQVVEGWYGQRFPKPLARTREYIDIVRQVWARESPVTSAGPHYRLP
Rv3520c|M.tuberculosis_H37Rv GVSGPQVVEGWYGQRFPKPLARTREYIDIVRQVWARESPVTSAGPHYRLP
MLBr_00348|M.leprae_Br4923 GVSGPQVVEGWYGQPFPKPLSRTREYIDIVRQVWAREAPVVSDGQHYPLP
*.*****:**::* : *:.**** ::* *:** ** : * :: :*
MUL_4943|M.ulcerans_Agy99 LPAGQGTGLGKPLKIINQPVQKRIPILLAALGPKNVELAAEAAEGWQPIF
MMAR_0154|M.marinum_M LPAGQGTGLGKPLKIINQPVRERIPILLAALGPKNVELAAEVAEGWQPIF
MAV_0137|M.avium_104 LPADRGTGLGKPLKLINSPVRERIPILVAALGPKNVEMAAEIAEGWQPIF
MSMEG_6602|M.smegmatis_MC2_155 LPADQGTGLGKALKLINQPVRERIPVLLAALGPKNVELAAEIAEGWQPIF
MAB_3748|M.abscessus_ATCC_1997 LPPEQGTGLGKALKLINHPVRDRIPVLIAALGPKNVALTAEIAEGWQPIF
Mflv_4941|M.gilvum_PYR-GCK LPAERGTGLGKPLKLINHPVRENIPITIAALGPKNVELTAEIAEGWQPVF
Mvan_1478|M.vanbaalenii_PYR-1 LPSDKGTGLGKPLKLINHPVRERIPITIAALGPKNVELTAEIAEGWQPVF
TH_1703|M.thermoresistible__bu LPPDRGTGLGKPLKLINHPVREHIPIFIAALGPKNVQLTAEIADGWQPVF
Mb3550c|M.bovis_AF2122/97 LTGEGTTGLGKALKPITHPLRADIPIMLGAEGPKNVALAAEICDGWLPIF
Rv3520c|M.tuberculosis_H37Rv LTGEGTTGLGKALKPITHPLRADIPIMLGAEGPKNVALAAEICDGWLPIF
MLBr_00348|M.leprae_Br4923 LTGAGATGLGKALKPITHPLRADIPIMLGAEGPKNVALAAEICDGWLPIF
*. *****.** *. *:: **: :.* ***** ::** .:** *:*
MUL_4943|M.ulcerans_Agy99 YLPEKAHDVWGQALAAGRAKRDPALG--ELDVYAGPVLAIGDNVESLREF
MMAR_0154|M.marinum_M YLPEKAHDVWGQALAAGRAKRDPALG--ELDVYAGPVLAIGDNVESLREF
MAV_0137|M.avium_104 YLPEKARDVWGESLAAGRAKRAAELG--ELEIYAGPVLAIGENTEPLREF
MSMEG_6602|M.smegmatis_MC2_155 FLPEKAATVWGDALKAGKTKRDPALG--ELNVYAGPVLAIGDNVEPLREF
MAB_3748|M.abscessus_ATCC_1997 FLPEKANDVWGAALAEGKAKRDPALG--DLEVYAGPALAIGDDVEALYGF
Mflv_4941|M.gilvum_PYR-GCK FYPEKADEVWGDALRAGASRRDPALG--PLDVMVSATLAIGDDVDDRLSW
Mvan_1478|M.vanbaalenii_PYR-1 FYPEKADDVWGDALRAGASRRDPALG--PLDTMVSASLAIGDDVDDRLAW
TH_1703|M.thermoresistible__bu VDPERIDSVWGDALRAGRAKRDPTLG--PLDVMGTPPLAIGDDVEDRIAW
Mb3550c|M.bovis_AF2122/97 YSP-RMAGMYNEWLDEGFARPGARRSREDFEICATAQVVITDDRAAAFAG
Rv3520c|M.tuberculosis_H37Rv YSP-RMAGMYNEWLDEGFARPGARRSREDFEICATAQVVITDDRAAAFAG
MLBr_00348|M.leprae_Br4923 FSP-RMADMYNEWLDEGFARTGARRSREDFEICASAQIVVTEDRAAAFAG
* : ::. * * :: . . :: . :.: ::
MUL_4943|M.ulcerans_Agy99 VKPHLALYIGGMGAKGKNFYHMLATRYGYGAEADRIQQLYLAGDKDGAAK
MMAR_0154|M.marinum_M VKPHLALYIGGMGAKGKNFYHMLATRYGYGAEADRIQQLYLAGDKDGAAK
MAV_0137|M.avium_104 VKPHLALYIGGMGAKGKNFYHALATKYGYGPQADRIQELYLAGDKDGAAK
MSMEG_6602|M.smegmatis_MC2_155 VKPHLALYIGGMGAKGKNFYHSLATSYGYGAEADRIQELYLAGEKDAAAK
MAB_3748|M.abscessus_ATCC_1997 VKPSLALYIGGMGAKGKNFYHNLATAYGFGKEADTIQELYLSGKKAEATD
Mflv_4941|M.gilvum_PYR-GCK GKPQLALYIGGMGARGKNFYHNLATRYGFGEVADHIQDLYLAGKKAEAID
Mvan_1478|M.vanbaalenii_PYR-1 AKPQLALYIGGMGARGKNFYHNLATRYGFGEVADHIQDLYLAGKKAEAID
TH_1703|M.thermoresistible__bu VKPMLALYLGGMGARDKNFYHNLATRYGFGAEADRIQELYLAGRKEEAAA
Mb3550c|M.bovis_AF2122/97 IKPFLALYMGGMGAEETNFHADVYRRMGYTQVVDEVTKLFRSGRKDEAAE
Rv3520c|M.tuberculosis_H37Rv IKPFLALYMGGMGAEETNFHADVYRRMGYTQVVDEVTKLFRSGRKDEAAE
MLBr_00348|M.leprae_Br4923 IKPFLALYMGGMGAEGTNFHADVYRRMGYAEVVDEVTALFRSNQKDKAAE
** ****:*****. .**: : *: .* : *: :. * *
MUL_4943|M.ulcerans_Agy99 VVPDELVRDVNLIGTPGFVRDRLAAFRESGVTTLNVAPIAETAAERVKLI
MMAR_0154|M.marinum_M VVPDELVRDVNLIGTPGFVRDRLAAFRESGVTTLNVAPIAETAAERVKLI
MAV_0137|M.avium_104 VVPDDLVRDVNLIGSREFVKERVAAFREAGVTTLNVAPMASTPAERVKLI
MSMEG_6602|M.smegmatis_MC2_155 AVPDELVRDVNLIGSAGFVRERIAAYREAGVTTLNVVPIGETAADRVRLI
MAB_3748|M.abscessus_ATCC_1997 AVPDELVKNISLIGPKSFVAERVAAFKEAGVTTLNVAPLAADAAGRIKHV
Mflv_4941|M.gilvum_PYR-GCK AVPDELVRQVSLVGPAGFVKERLAAFAEAGVTTLLVHPMGVDAAERVRFV
Mvan_1478|M.vanbaalenii_PYR-1 AVPDELVRQVSLVGPAGFVKERLAAYAEAGVTTMLVHPLSGDRRETVKFV
TH_1703|M.thermoresistible__bu AVPDDLVRSVSLIGPAGLVAERVAMFAEAGVTTLLVTPLATDTAEAVRFI
Mb3550c|M.bovis_AF2122/97 IIPDELVDDAVIVGDIDHVRKQMAVWEAAGVTMMVVT------AGSAEQV
Rv3520c|M.tuberculosis_H37Rv IIPDELVDDAVIVGDIDHVRKQMAVWEAAGVTMMVVT------AGSAEQV
MLBr_00348|M.leprae_Br4923 IIPNELVDSTMIVGDVDYVCKQIAAWSAAGVTMMMVS------ASSVEQV
:*::** . ::* * .::* : :*** : * . :
MUL_4943|M.ulcerans_Agy99 DTLRQLVD
MMAR_0154|M.marinum_M DTLRQLVD
MAV_0137|M.avium_104 EALRQLV-
MSMEG_6602|M.smegmatis_MC2_155 EQLRELV-
MAB_3748|M.abscessus_ATCC_1997 EALRELL-
Mflv_4941|M.gilvum_PYR-GCK EQLQGLL-
Mvan_1478|M.vanbaalenii_PYR-1 EELQNLLP
TH_1703|M.thermoresistible__bu EQLRTMVA
Mb3550c|M.bovis_AF2122/97 RDLAALV-
Rv3520c|M.tuberculosis_H37Rv RDLAALV-
MLBr_00348|M.leprae_Br4923 RDLAGLF-
* :.