For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TGPARPWRVLIVVALLLVYTAPSADAVGPPAVDERLLPAPGPVAPAHPTVQHDECVTAPTSPVDVPGRPH GIDLPSVWALTRGAGQTVAVIDTGVARHRRLPHLIAGGDYVSGQDGTSDCDGHGTIVAGIIGASADPADT GGFAGIAPDVSLLSIRQSSNRFRAVDDPGGTGVGDVETLATAVRTAADLGATVLNISTVACLPVDAGLDD RALGAALAYAVDVKNVVVVTAAGNVGGAGQCTQHNPRRPADAGAVTMVASPAWYDDYVLTVGSIGSDGIA SEFSLAGPWVDVAAPGESVVSLAPAGDGLVDVAPSGLPISGTSYAVPVVSAIAALVRARSPGLTARQVMR VIEDTAHPPPSGWDPVVGHGVVDALAAVTATGSADPVSAGVAPVGPVPPRPSESVSGRTALHAALGCLAL TACVAVVSTAVRRSGRPGSVAQD*
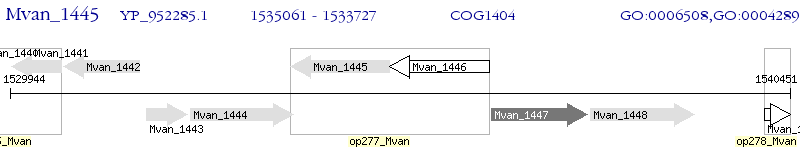
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_1445 | - | - | 100% (444) | peptidase S8 and S53, subtilisin, kexin, sedolisin |
| M. vanbaalenii PYR-1 | Mvan_0421 | - | 1e-81 | 42.64% (455) | peptidase S8 and S53, subtilisin, kexin, sedolisin |
| M. vanbaalenii PYR-1 | Mvan_0092 | - | 5e-69 | 38.89% (450) | peptidase S8 and S53, subtilisin, kexin, sedolisin |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3479 | - | 1e-130 | 56.95% (453) | secreted serine protease |
| M. gilvum PYR-GCK | Mflv_4961 | - | 1e-169 | 69.66% (435) | peptidase S8 and S53, subtilisin, kexin, sedolisin |
| M. tuberculosis H37Rv | Rv3449 | mycP4 | 1e-130 | 56.95% (453) | membrane-anchored mycosin |
| M. leprae Br4923 | MLBr_02528 | - | 2e-79 | 39.05% (461) | putative protease |
| M. abscessus ATCC 19977 | MAB_3758 | - | 1e-112 | 53.72% (443) | putative protease |
| M. marinum M | MMAR_1100 | mycP4 | 1e-121 | 55.84% (428) | membrane-anchored serine protease (mycosin), MycP4 |
| M. avium 104 | MAV_4393 | - | 1e-126 | 57.17% (446) | peptidase |
| M. smegmatis MC2 155 | MSMEG_1533 | - | 1e-123 | 58.16% (435) | subtilase family protein |
| M. thermoresistible (build 8) | TH_3410 | - | 1e-124 | 57.80% (436) | PUTATIVE peptidase S8 and S53, subtilisin, kexin, sedolisin |
| M. ulcerans Agy99 | MUL_0858 | mycP4 | 1e-120 | 55.61% (428) | membrane-anchored serine protease (mycosin), MycP4 |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_1445|M.vanbaalenii_PYR-1 ----------MTGPARPWRVLIVVALLLVYTAPSADAVGPPAVDERLLPA
Mflv_4961|M.gilvum_PYR-GCK ----------MT---RTWRVAAVTAIALMHTTPTAAAIGPPPVDTLRLPT
MSMEG_1533|M.smegmatis_MC2_155 --------MMVAQRVTATLMTVAVVSLTLVPAPPAAAVTPPTVDDAYLPP
TH_3410|M.thermoresistible__bu -------VVRSAPARLARGLAIAVTASLVGAAPGAHAVTPPPIDHARLPH
Mb3479|M.bovis_AF2122/97 ---------MTTSRTLRLLVVSALATLSGLGTPVAHAVSPPPIDERWLPE
Rv3449|M.tuberculosis_H37Rv ---------MTTSRTLRLLVVSALATLSGLGTPVAHAVSPPPIDERWLPE
MMAR_1100|M.marinum_M ----------MTSRFGHLLAACALAAPCWCAMPAAHAVTPPPVDERWLPQ
MUL_0858|M.ulcerans_Agy99 ----------MTSRFGHLLAACALAAPCWCAMPAAHAVTPPPVDERWLPQ
MAV_4393|M.avium_104 ---------------MRLLAVSALTMLPVFETPPAHAVSPPAVDDKWLPK
MAB_3758|M.abscessus_ATCC_1997 MSLGVARRIATAGAVVMLAMSSQLATGSMHVMATAHAFAPLPVDPSMLPG
MLBr_02528|M.leprae_Br4923 ----MMRSELRIGLAGIATTLVVFQATLLRGCPWALAISPPVVDPGVPPP
. * *. * :* *
Mvan_1445|M.vanbaalenii_PYR-1 PGPVAPAHPTVQHDECVTAPTSPV------DVPGRPHGIDLPSVWALTRG
Mflv_4961|M.gilvum_PYR-GCK ADPPAPAQPTVQFTDCVAAPPAAG------SAPGQLAGMDLPTVWALTRG
MSMEG_1533|M.smegmatis_MC2_155 AAPPAPPHPTEQRRPCFD-NRGRD----DDETRS--LSPALEPVWRLTRG
TH_3410|M.thermoresistible__bu AGPAAPPQRTEQQGPCLRPAQRRT----GDAVLSGFTAADLAAVWRLTRG
Mb3479|M.bovis_AF2122/97 SALPAPPRPTVQREVCTEVTAESGRAFGRAERSAQLA--DLDQVWRLTRG
Rv3449|M.tuberculosis_H37Rv SALPAPPRPTVQREVCTEVTAESGRAFGRAERSAQLA--DLDQVWRLTRG
MMAR_1100|M.marinum_M PAPPGPPSRTRQREVCAAATWAPE----RPEAAAQLAGLDLPEAWRLTRG
MUL_0858|M.ulcerans_Agy99 PAPPGPPSRTRQREVCAAATWAPE----RPEAAAQLAGLDLPEAWRLTRG
MAV_4393|M.avium_104 PARPAPPWPTAQREVCATLTADSG-----PGRRGPAGGSDLSAVWQLSRG
MAB_3758|M.abscessus_ATCC_1997 PRPPAPIEPTEQKELCRFPGIDPKR--DLNKVPTQQRGLDLPLVWQLSRG
MLBr_02528|M.leprae_Br4923 NGAPGPVQIMQQRGSCITSGVIPG--TDLGIVTSSQSMLNLPAAWQFSRG
.* * * * .* ::**
Mvan_1445|M.vanbaalenii_PYR-1 AGQTVAVIDTGVARHRRLPHLIAGGDYVS-GQDGTSDCDGHGTIVAGIIG
Mflv_4961|M.gilvum_PYR-GCK AGQTVAVIDTGTARHRRLPHLVPGGDYVS-HRDGTDDCDGHGTLVAGIIG
MSMEG_1533|M.smegmatis_MC2_155 RGQVVAVIDTGVARHRWLPRLVGGGDYVS-NGDGTADCDGHGTIVAGIIG
TH_3410|M.thermoresistible__bu GGQAVAVIDTGVARHRLLPRVIPGGDFVH-RGDGTDDCDGHGTIVAGLIA
Mb3479|M.bovis_AF2122/97 AGQRVAVIDTGVARHRRLPKVVAGGDYVF-TGDGTADCDAHGTLVAGIIA
Rv3449|M.tuberculosis_H37Rv AGQRVAVIDTGVARHRRLPKVVAGGDYVF-TGDGTADCDAHGTLVAGIIA
MMAR_1100|M.marinum_M SGQRVAVIDTGVAHHRRLAHLVAGGDYVF-TGDGTQDCDAHGTIVAGIIA
MUL_0858|M.ulcerans_Agy99 SGQRVAVIDTGVAHHRRLAHLVAGGDYVF-TGDGTQDCDAHGTIVAGIIA
MAV_4393|M.avium_104 TGQRVAVIDTGVARHRRLPAVVPGGDYVS-TGDGIQDCDAHGTLVAGIIA
MAB_3758|M.abscessus_ATCC_1997 AGQTVAVIDTGVARSPRLPPLIPGGDYVFEGGNGTQDCDAHGTIVAGIIA
MLBr_02528|M.leprae_Br4923 EGQLVAIVDTGVRPGPRLPNVEAGGDYVE-SSDGLTDCDGHGTLVAGIVA
** **::***. *. : ***:* :* ***.***:***::.
Mvan_1445|M.vanbaalenii_PYR-1 ASADPADTGGFAGIAPDVSLLSIRQSSNRFRAVDDPGG--------TGVG
Mflv_4961|M.gilvum_PYR-GCK ASPDAADPGGFTGIAPDVSLLSIRQSSNKFRAVDDPGG--------SGVG
MSMEG_1533|M.smegmatis_MC2_155 GAATP---DGFSGVAPEATILAIRQSSNRF--ADDAGG--------SGVG
TH_3410|M.thermoresistible__bu AAPGDG-RAGFSGIAPESSIIAIRQSSNKFRATDQPSG--------TGYG
Mb3479|M.bovis_AF2122/97 AAPDAQ-SDNFSGVAPDVTLISIRQSSSKFAPVGDPSS--------TGVG
Rv3449|M.tuberculosis_H37Rv AAPDAQ-SDNFSGVAPDVTLISIRQSSSKFAPVGDPSS--------TGVG
MMAR_1100|M.marinum_M AAPDSA-TDQFSGVAPDATVIGIRQSSAKFSPVGNRSS--------TGVG
MUL_0858|M.ulcerans_Agy99 AAPDSA-TDQFSGVAPDATVIGIRQSSAKFSPVGNRSS--------TGVG
MAV_4393|M.avium_104 AAADPK-SDEFSGIAPEATLISIRQSSAKFGPAGNRSG--------GGVG
MAB_3758|M.abscessus_ATCC_1997 AAPDPTGASAFSGIAPDVSLITIRQSSAKFAVKNKGGNDRDDPSKISGVG
MLBr_02528|M.leprae_Br4923 GQPAPSGEDGFSGVAPAARLLSIRVTSAKFSLRSLGGD----PQLKQASL
. . *:*:** :: ** :* :* . .. .
Mvan_1445|M.vanbaalenii_PYR-1 DVETLATAVRTAADLGATVLNISTVACLPVDAGLD-------DRALGAAL
Mflv_4961|M.gilvum_PYR-GCK DVETLAAAVRTAADLGATVINISTVACLPVAAGLD-------DRTLGAAL
MSMEG_1533|M.smegmatis_MC2_155 DVGTLAKAVRTAADLGATVINISSVACVDADPDTDTADAGFDDRPLGAAL
TH_3410|M.thermoresistible__bu DVDTLAMAIRTAADLGASVINVSSVACLAATEPPD-------DRALGAAL
Mb3479|M.bovis_AF2122/97 DVDTMAKAVRTAADLGASVINISSIACVPAAAAPD-------DRALGAAL
Rv3449|M.tuberculosis_H37Rv DVDTMAKAVRTAADLGASVINISSIACVPAAAAPD-------DRALGAAL
MMAR_1100|M.marinum_M DVDTMARAVRTAADLGASVINISTIACVAAESPPD-------DRALGAAL
MUL_0858|M.ulcerans_Agy99 DVDTMARAVRTAADLGASVINISTIACVAAESPPD-------DRALGAAL
MAV_4393|M.avium_104 DVDTLAKAVRTAADLGASVINISSVACAPVVSAPD-------DRALGAAL
MAB_3758|M.abscessus_ATCC_1997 NVDTLAMAVRTAADMGATVINISEVACASAKQFLD-------DRPLGAAL
MLBr_02528|M.leprae_Br4923 EVTRLSRAIVHAADLGARVINISAITCLPADRSID-------QSGLGAAI
:* :: *: ***:** *:*:* ::* . * : ****:
Mvan_1445|M.vanbaalenii_PYR-1 AYAVDVKNVVVVTAAGNVGGAGQCT-----QHNPRR-------PADAGAV
Mflv_4961|M.gilvum_PYR-GCK SYAVEAKNVVVVTAAGNVGGPGQCQ-----DPNPRD-------VGAADQV
MSMEG_1533|M.smegmatis_MC2_155 AYAVDVKNVVVVAAAGNVG-AGQCR-----HQNPLD-------APGWDDV
TH_3410|M.thermoresistible__bu AYAVDVRNAVVVSAAGNVGGAGQCP-----HQNPVTG-----RRPDWATV
Mb3479|M.bovis_AF2122/97 AYAVDVKNAVIVAAAGNTGGAAQCP-----PQAPGV---------TRDSV
Rv3449|M.tuberculosis_H37Rv AYAVDVKNAVIVAAAGNTGGAAQCP-----PQAPGV---------TRDSV
MMAR_1100|M.marinum_M AYAVDVKNAVIVAAAGNTGGAAQCP-----SQRPET---------SRDTA
MUL_0858|M.ulcerans_Agy99 AYAVDVKNAVIVAAAGNTGGAAQCP-----SQRPET---------SRDTA
MAV_4393|M.avium_104 AYAVDVKNAVVVAAAGNTGGPGQCP-----PQRPDA---------TWQTI
MAB_3758|M.abscessus_ATCC_1997 QYAVDVKNVVVVVAAGNVG-DGQCK-----SQNPPPDPARP-NTPDWDSV
MLBr_02528|M.leprae_Br4923 RYAAVDKDALLVAPAGNAGSTGTVGGNMNCESNPLTDLGRPDDPRNWASV
**. ::.::* .***.* . *
Mvan_1445|M.vanbaalenii_PYR-1 TMVASPAWYDDYVLTVGSIGSDGIASEFSLAGPWVDVAAPGESVVSLAPA
Mflv_4961|M.gilvum_PYR-GCK RTVASPAWYDDYVLTVGSVGTDGSPSEFSLAGPWVDVAATGEGVVSLAAT
MSMEG_1533|M.smegmatis_MC2_155 QSVTSPGWYDDLVLCVGSVAADGTASSFSLAGPWVDIAAPGEQVVSLHPD
TH_3410|M.thermoresistible__bu RAVVTPAWYDDLVLTVGSVNAEGHPSDFTLAGPWVDVAAPGEDVLSLDPD
Mb3479|M.bovis_AF2122/97 TVAVSPAWYDDYVLTVGSVNAQGEPSAFTLAGPWVDVAATGEAVTSLSPF
Rv3449|M.tuberculosis_H37Rv TVAVSPAWYDDYVLTVGSVNAQGEPSAFTLAGPWVDVAATGEAVTSLSPF
MMAR_1100|M.marinum_M TVVVSPAWYDDYVLTVGSVNANGEPSAFTLPSPWVDVAAGGENVTSLNPV
MUL_0858|M.ulcerans_Agy99 TVVVSPAWYDNYVLTVGSVNANGEPSAFTLPSPWVDVAAGGENVTSLNPV
MAV_4393|M.avium_104 TVAVSPAWYDDYVLTVGSVNADGAPSTFSLAGPWVDVAAPGEAVTSLAP-
MAB_3758|M.abscessus_ATCC_1997 LETVSPAWYNDLVLTVGSIGPDGQPSKFSLGGPWVDVSAPGEDIVSLDPN
MLBr_02528|M.leprae_Br4923 TSVSIPSWWQPYVLSVASLTPSGRPSKFTMAGPWVGIAAPGENIVSVSNR
. *.*:: ** *.*: ..* .* *:: .***.::* ** : *:
Mvan_1445|M.vanbaalenii_PYR-1 GDG-LVDVAP----SGLPISGTSYAVPVVSAIAALVRARSPGLTARQVMR
Mflv_4961|M.gilvum_PYR-GCK GDG-LADRSP----SDAALTGTSYAVPVVSAVAALVRARFPEMTARQVMQ
MSMEG_1533|M.smegmatis_MC2_155 GDG-LIASL----GADKPISGTSYAAPFVAGVAALVRSRFPHLSAREVMR
TH_3410|M.thermoresistible__bu GEG-LVDALPGP-GGDTPIAGTSYAAPLVAGLAALVRARWPQLTARQVMQ
Mb3479|M.bovis_AF2122/97 GDG-TVNRLGGQ-HGSIPISGTSYAAPVVSGLAALIRARFPTLTARQVMQ
Rv3449|M.tuberculosis_H37Rv GDG-TVNRLGGQ-HGSIPISGTSYAAPVVSGLAALIRARFPTLTARQVMQ
MMAR_1100|M.marinum_M GDG-TVNGLDDH-GGFRPLSGTSYAAPVVSGLAALIRARFPALTARQVMA
MUL_0858|M.ulcerans_Agy99 GDG-TVNGLDDH-GGFRPLSGTSYAAPVVSGLAALIRARFPALTARQVMA
MAV_4393|M.avium_104 ----------------EPVSGTSYAAPVVSGLAALIRARFPALTARQVMQ
MAB_3758|M.abscessus_ATCC_1997 RDA-LTNAYMGD-QGPVPIQGTSFSAPVVSGVVALVRSRFPQLSAREVMD
MLBr_02528|M.leprae_Br4923 DSGDLANALPNDRDQQIALNGTSYAAGYVSGVAALVRSKYPELTAKQVMR
.: ***::. *:.:.**:*:: * ::*::**
Mvan_1445|M.vanbaalenii_PYR-1 VIEDTAHPPPSGWDPVVGHGVVDALAAVTAT--------GSADPVSAGVA
Mflv_4961|M.gilvum_PYR-GCK RIEDTARPSSAEWDPVVGHGVVDALAAVSGP--------GSHPAALPAAP
MSMEG_1533|M.smegmatis_MC2_155 RIEATAVRPPRGWDAAVGSGVVDPLAAVSGD-----APALAPPARNTAMV
TH_3410|M.thermoresistible__bu RIRDTARTPASGWNPLVGHGVIDPLAAVSGDP----APGRPAPAHLTRTL
Mb3479|M.bovis_AF2122/97 RIESTAHHPPAGWDPLVGNGTVDALAAVSSDSIPQAGTATSDPAPVAVPV
Rv3449|M.tuberculosis_H37Rv RIESTAHHPPAGWDPLVGNGTVDALAAVSSDSIPQAGTATSDPAPVAVPV
MMAR_1100|M.marinum_M RIISTAHHPPHGWDPFVGNGTVDVLAAVSSD--PAVQPAPAQPGSPTPAA
MUL_0858|M.ulcerans_Agy99 RIISTAHHPPHGWDPFVGNGTVDVLAAVSSD--PAVQPAPAQPGSPTPAA
MAV_4393|M.avium_104 RIEATAHHPPAGWDPLVGHGTVDPLAAVSAE------TGAPPPAPRAAAV
MAB_3758|M.abscessus_ATCC_1997 RIKNTAHRPPGGWDPYLGFGVVDALAAVSDD-----LGTTSDLARANAAK
MLBr_02528|M.leprae_Br4923 RITATAHTGARTLSNVVGSGTLDSVAALTWD------LPATNDSPPVPVK
* ** . . :* *.:* :**:: .
Mvan_1445|M.vanbaalenii_PYR-1 PVGPVPPRPSES-VSGRTALHAALGCLALTACVAVVS----------TAV
Mflv_4961|M.gilvum_PYR-GCK PVDAPTPAPARVQRQGATAVRGALICVALTVCVALVS----------AVI
MSMEG_1533|M.smegmatis_MC2_155 APPAPSPEDTG---PRRIAFGGGALCVAVA-----------LATVMLSRR
TH_3410|M.thermoresistible__bu PAPTTPAADPA---PRRIAFGGALLCLAAXXPLRYRMPGFFTNVISISSR
Mb3479|M.bovis_AF2122/97 PRRSTPGPSDR--RALHTAFAGAAICLLALMATLAT----------ASRR
Rv3449|M.tuberculosis_H37Rv PRRSTPGPSDR--RALHTAFAGAAICLLALMATLAT----------ASRR
MMAR_1100|M.marinum_M PVTTPPAQPDR--RARITALSGSAICLMGLLAALASRA---FGGRSGSRH
MUL_0858|M.ulcerans_Agy99 PVTTPPTQPDR--RARITALSGSAICLMGLLGALASRA---FGGRSGSRH
MAV_4393|M.avium_104 PIAAPPPAPARDSRAPDVAVRGAAFCLVALTLAAAAG--------AVRAR
MAB_3758|M.abscessus_ATCC_1997 KLVLPPAPPPPDPRPRRVALIGIAVCAAFLAAALAMLAP--------VRR
MLBr_02528|M.leprae_Br4923 LVTVPPPPAPKDNTPRNVAFAGTATLVMVVAAVVGT-----------ATI
. *. .
Mvan_1445|M.vanbaalenii_PYR-1 RRSGRPGSVAQD-------------------------
Mflv_4961|M.gilvum_PYR-GCK ARSRRHG------------------------------
MSMEG_1533|M.smegmatis_MC2_155 ---GRQGITGD--------------------------
TH_3410|M.thermoresistible__bu TLGGRSAGSGGGVSLMCFIATVNAPSPVNGRRPDTAS
Mb3479|M.bovis_AF2122/97 LRPGRNGIAGD--------------------------
Rv3449|M.tuberculosis_H37Rv LRPGRNGIAGD--------------------------
MMAR_1100|M.marinum_M RRIHRDDVVGD--------------------------
MUL_0858|M.ulcerans_Agy99 RRIHRDDVVGD--------------------------
MAV_4393|M.avium_104 LRRTRDGVAGD--------------------------
MAB_3758|M.abscessus_ATCC_1997 LRQSREHPDE---------------------------
MLBr_02528|M.leprae_Br4923 AARRREGTTP---------------------------
*