For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTPKRNPRRADLVLSGGGVKGIGLVGAVGALRDAGYALERISGTSAGSLVGAVVAAATQAGGISADQLLE LAVNVPYRRFRDSGPIERIPLLGTAWGLLRGTGIYRGDFAHQWIRGELKNLGVSTFGDLAVDDDNLLPER RYRLVVTVADLTTGQLVRLPWDYKRLYGLDPDEQSVADAVRASMSIPFLFRPSTLKDAAGQRCTLVDGGV LSNFPIDSFDRLDGAEPRWPTFGVTVVPYLPAPTTDQLIPGLRLPWDGPTFLESLITTMLVGHDQAHLSQ PWVKVRAIQVDSTDVGVLDFDITPSEADALYDKGYAAASEFLASWDWPAYLQRFRRTHYVT
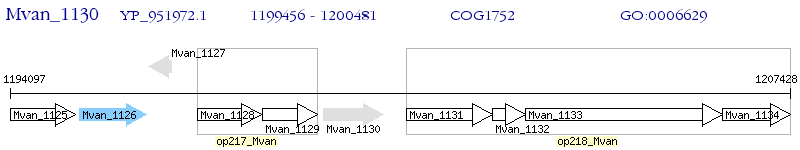
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_1130 | - | - | 100% (341) | patatin |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2063c | - | 3e-66 | 44.62% (325) | transmembrane protein |
| M. gilvum PYR-GCK | Mflv_2030 | - | 1e-07 | 28.63% (248) | patatin |
| M. tuberculosis H37Rv | Rv2037c | - | 3e-65 | 44.55% (321) | transmembrane protein |
| M. leprae Br4923 | MLBr_01423 | - | 2e-62 | 41.98% (324) | hypothetical protein MLBr_01423 |
| M. abscessus ATCC 19977 | MAB_4751 | - | 4e-08 | 27.86% (201) | hypothetical protein MAB_4751 |
| M. marinum M | MMAR_0961 | - | 1e-113 | 60.60% (335) | hypothetical protein MMAR_0961 |
| M. avium 104 | MAV_4537 | - | 1e-118 | 63.24% (340) | phospholipase, patatin family protein |
| M. smegmatis MC2 155 | MSMEG_5284 | - | 3e-06 | 28.37% (208) | patatin |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_2251 | - | 4e-67 | 45.73% (328) | hypothetical protein MUL_2251 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0961|M.marinum_M ------MKHGSKPVDLVLSGGGVKFMGLVGAIARLMDDGYQPYRVSGVSA
MAV_4537|M.avium_104 ---MTSKMEPAKPVDLVLSGGGVKFIGLVGAIVALMDAGYSIKRVSGVSA
Mvan_1130|M.vanbaalenii_PYR-1 ---MTPKRNPRR-ADLVLSGGGVKGIGLVGAVGALRDAGYALERISGTSA
Mb2063c|M.bovis_AF2122/97 -----MALVSTARVDLVCEGGGVRGIGLVGAVDALADAGYRFPRVAGSSA
Rv2037c|M.tuberculosis_H37Rv -----MALVSTARVDLVCEGGGVRGIGLVGAVDALADAGYRFPRVAGSSA
MUL_2251|M.ulcerans_Agy99 --------MNAARVDLVCEGGGVRGIGLVGAVDALAEAGYEFPRVAGSSV
MLBr_01423|M.leprae_Br4923 --MLYLVIVSVVHADLVCQGGGIRGIGLVGAVDALATAGYRFPRVAGTSA
Mflv_2030|M.gilvum_PYR-GCK -----------MRVALALGSGGARGYAHIGVLNELRERGHEVVGIAGSSM
MSMEG_5284|M.smegmatis_MC2_155 MTHNES-----MRVALALGSGSARGYAHIGVINELRARGYEIVGISGSSM
MAB_4751|M.abscessus_ATCC_1997 MTGEASTGVPGTKIALALGAGGARGYAHIGVIQELHERGFEISGISGSSM
*. .*. : . :*.: * *. ::* *
MMAR_0961|M.marinum_M GSVVAVIAAAGTQGNQLSSDQIRELALSVPLSTWR-DAAP---VPILGSA
MAV_4537|M.avium_104 GSVVAAILAAGCHAGELTGAQVKELAFSVPLHKWR-DAGP---VPYLGAA
Mvan_1130|M.vanbaalenii_PYR-1 GSLVGAVVAAATQAGGISADQLLELAVNVPYRRFR-DSGPIERIPLLGTA
Mb2063c|M.bovis_AF2122/97 GAIVASLVAALQTAGEPVT-RLAEMMRSIDYPKFL-DRNLIGHVPLIGGG
Rv2037c|M.tuberculosis_H37Rv GAIVASLVAALQTAGEPVT-RLAEMMRSIDYPKFL-DRNLIGHVPLIGGG
MUL_2251|M.ulcerans_Agy99 GAIVAAMVAALQTAGEPLS-RLAEIMRTLDYRKFL-DRNLIGHIPLIGGG
MLBr_01423|M.leprae_Br4923 GALVASLIAALQTAGEPLT-RLAEVMRTIDYRKFF-DRSLIGRVPLIGGG
Mflv_2030|M.gilvum_PYR-GCK GALVGGLEAAGKTD------EFTLWATSLTQRAVLRLLDPSISSPGVLRA
MSMEG_5284|M.smegmatis_MC2_155 GALVGGLYAAGKLD------EFAEWACTLTQRAVLRLLDPSLSAAGILRA
MAB_4751|M.abscessus_ATCC_1997 GALVGGLEAAGALD------EFALWATSLTQRAVLRLLDPTWTSPGFFRA
*::*. : ** .. .: . . .
MMAR_0961|M.marinum_M WGLVRDSAMYRGDAAYDWIRSELENLGVTTFGDLALDDDELPVEERYRVA
MAV_4537|M.avium_104 WGLARDTSMYRGDVAHDWIRSELKNFGVSTFGDLVFDGDDLPDERRRRLV
Mvan_1130|M.vanbaalenii_PYR-1 WGLLRGTGIYRGDFAHQWIRGELKNLGVSTFGDLAVDDDNLLPERRYRLV
Mb2063c|M.bovis_AF2122/97 LSLLLSDGVYRGAYLEQLLGGLLADLGVHTFGDLRTGEA--PEQFAWSLV
Rv2037c|M.tuberculosis_H37Rv LSLLLSDGVYRGAYLEQLLGGLLADLGVHTFGDLRTGEA--PEQFAWSLV
MUL_2251|M.ulcerans_Agy99 LSLLVSDGVYRGAYLEQLLAGLLADLGVHTFGDLRTGEQ--PEQFAWSLV
MLBr_01423|M.leprae_Br4923 LSLLVSDGVYQGAYLEEWLTSLLGDLGVYTFGDLRTGEE--PEQFAWSLV
Mflv_2030|M.gilvum_PYR-GCK EKILESVRDILGEVTIEDLPIPYTSVATDLITGKSVWMQRGRVDDAIRAS
MSMEG_5284|M.smegmatis_MC2_155 EKILDAVRDILGDVTIEELPIPYTAVATDLITGKAVWLQRGRVDTAIRAS
MAB_4751|M.abscessus_ATCC_1997 EKILDVVRELLGDVAIEDLPIPFTAVATDLIAGKSVWLQNGSLASAIRAS
: * : : ... : .
MMAR_0961|M.marinum_M ITVADVTRAQLVRLPWDYRRVYGVDPDEQSVASAVRASMAIPFIYPPVTL
MAV_4537|M.avium_104 VTVADVTAAQLVRLPWDYRRLYGLDPDEQPVADAVRASMAIPFFYRPVKL
Mvan_1130|M.vanbaalenii_PYR-1 VTVADLTTGQLVRLPWDYKRLYGLDPDEQSVADAVRASMSIPFLFRPSTL
Mb2063c|M.bovis_AF2122/97 VTASDLSRRRLVRIPWDLD-SYGIHPDDFSVARAVHASSAIPFVFEPVRV
Rv2037c|M.tuberculosis_H37Rv VTASDLSRRRLVRIPWDLD-SYGIHPDDFSVARAVHASSAIPFVFEPVRV
MUL_2251|M.ulcerans_Agy99 VTASDLSRRRLVRIPWDLD-SYGLDPDEFSVARAVHASSAIPFVFEPVRV
MLBr_01423|M.leprae_Br4923 VTASDLSRRRLVRIPWDLD-AYGINPDDFSVARAVHASSAIPYVFEPVQV
Mflv_2030|M.gilvum_PYR-GCK IAIPGLITPHVLGGRLLAD-GGILDPLPMAPIAAAPADITIAVSLSG-ED
MSMEG_5284|M.smegmatis_MC2_155 IAIPGVIAPHVHDGRLLGD-GGILDPLPMAPIASVNADLTIAVSVSGSET
MAB_4751|M.abscessus_ATCC_1997 IAIPGMISPHVLNGRVLAD-GGVLDLPAVAPLAGVHAELRVAVTLSGGDE
:: ..: :: :. . .. *. :.
MMAR_0961|M.marinum_M TSSDGLRSTLVDGG-VLSNFPVDSLDRTDTKPPRWPTFGITVIP-KLAEG
MAV_4537|M.avium_104 ARADGTACTLVDGG-VLSNFPIDTFDRPDGRAPRWPTFGITVMP-SPTEG
Mvan_1130|M.vanbaalenii_PYR-1 KDAAGQRCTLVDGG-VLSNFPIDSFDRLDGAEPRWPTFGVTVVPYLPAPT
Mb2063c|M.bovis_AF2122/97 RG-----ATWVDGG-LLSNFPVALFDRTDA-EPRWPTFGIRLSA---RPG
Rv2037c|M.tuberculosis_H37Rv RG-----ATWVDGG-LLSNFPVALFDRTDA-EPRWPTFGIRLSA---RPG
MUL_2251|M.ulcerans_Agy99 RG-----ATWVDGG-LLSNFPVALFDRADG-EPRWPTFGIRLSA---RPG
MLBr_01423|M.leprae_Br4923 AG-----ATWVDGG-LLSTFPVELFDRSDG-DARWPTFGIRLSA---RPG
Mflv_2030|M.gilvum_PYR-GCK P---AVDRRMQDLR----QPTTDWLSRMWRSSSSLLDTKAARSLLETPTG
MSMEG_5284|M.smegmatis_MC2_155 PPETASEAHSEPAEGGGTKPTTEWLNRMMRSTSAVLDTAAVRSVLDRPTS
MAB_4751|M.abscessus_ATCC_1997 RRAPAPPEPEPEPR--EPRVTTEWLNRLLRSTSALFST------------
. :.* .
MMAR_0961|M.marinum_M IEQAFPALRPLRFLQQSALLESLFTTMLVGHDQ-----AHLSQPWVSART
MAV_4537|M.avium_104 IGAVMPALKPLRFLPQTALLESLLITMLAGHDQ-----THLSQPWVAARA
Mvan_1130|M.vanbaalenii_PYR-1 TDQLIPGLRLP--WDGPTFLESLITTMLVGHDQ-----AHLSQPWVKVRA
Mb2063c|M.bovis_AF2122/97 IPPTRPVQGPVS------LGIAAIETLVSNQDN-----AYIDDPCTVRRT
Rv2037c|M.tuberculosis_H37Rv IPPTRPVQGPVS------LGIAAIETLVSNQDN-----AYIDDPCTVRRT
MUL_2251|M.ulcerans_Agy99 IPPTRPVHGPVS------LGIAAIETLVSNQDN-----AYIDDPCTVQRT
MLBr_01423|M.leprae_Br4923 IPPTHPVRGPVS------LGIAAIETLVSNQDN-----AYIDDSCTMLRT
Mflv_2030|M.gilvum_PYR-GCK RAVLGRF---VAADDEPIDAPPEVVKHDSADATGVPELPRLGSFEVMNRV
MSMEG_5284|M.smegmatis_MC2_155 RAVLSRFGVSVTESDDGTDASEDATVPESSAEP--AGVPRLGSFEVMYRT
MAB_4751|M.abscessus_ATCC_1997 ------------------DDPEVAGQPDP--------AAKLTSFEVMNRT
. : . . *.
MMAR_0961|M.marinum_M IAVASTDVGVLDFGIGRARLEELYQRGYAAAEAFLSTWDWADYLHRFRRF
MAV_4537|M.avium_104 IAVESTNVGVLDFDVPRSRLEELYDSGYAAAQAFLSTWDWQAYLDRFRYP
Mvan_1130|M.vanbaalenii_PYR-1 IQVDSTDVGVLDFDITPSEADALYDKGYAAASEFLASWDWPAYLQRFRRT
Mb2063c|M.bovis_AF2122/97 IFVPAHDVSPIDFDITAEQREALYQRGFQAGQKFLANWNYADYLADCGGP
Rv2037c|M.tuberculosis_H37Rv IFVPAHDVSPIDFDITAEQREALYQRGFQAGQKFLANWNYADCLADCGGP
MUL_2251|M.ulcerans_Agy99 IFVPADGVSPIDFDITSEQRDALYRRGLQAGRKFLQSWNYADYLTECGSP
MLBr_01423|M.leprae_Br4923 IFVPADEVSPIDFNITVEQRQALYQSGLQAGQQFLKTWNYTEYLTACGRP
Mflv_2030|M.gilvum_PYR-GCK IDIAQSALARHTLAAYPPDLLIEVPRTVCRSLDFHRAHEVIEIGQELASA
MSMEG_5284|M.smegmatis_MC2_155 IDIAQAALARHTLAAYPPDLLIEVPRTVCRSLEFNRAAEVIETGRRLAAE
MAB_4751|M.abscessus_ATCC_1997 IEVMQSQLARMQLAAHPPDVLIEVPRSVSRSLDFHRATEVIEVGRRCAAE
* : :. : . * :
MMAR_0961|M.marinum_M G-----------
MAV_4537|M.avium_104 GGRG--------
Mvan_1130|M.vanbaalenii_PYR-1 HYVT--------
Mb2063c|M.bovis_AF2122/97 FTPSL-------
Rv2037c|M.tuberculosis_H37Rv FTPSL-------
MUL_2251|M.ulcerans_Agy99 AAPSP-------
MLBr_01423|M.leprae_Br4923 VTRSAGL-----
Mflv_2030|M.gilvum_PYR-GCK ALEALDPEPSEA
MSMEG_5284|M.smegmatis_MC2_155 ALDS--------
MAB_4751|M.abscessus_ATCC_1997 ALDAYAHTN---