For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PDRHSVLRARTAVDVFAEVRFGLQRLDPPGIDMTTLTRLSAIAAMAAAAAVALAASAAAAPQDAVFLDRL QKVGITSSNPYATVHGGYDVCRQLDLGTPHSQIVGFVLSDNPDLDWDSAADYVVLANMTYCPPV*
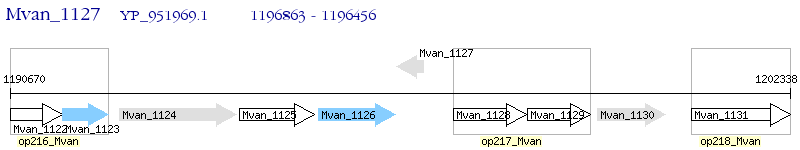
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_1127 | - | - | 100% (135) | hypothetical protein Mvan_1127 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1302c | - | 3e-09 | 34.41% (93) | hypothetical protein Mb1302c |
| M. gilvum PYR-GCK | Mflv_5185 | - | 3e-37 | 73.20% (97) | hypothetical protein Mflv_5185 |
| M. tuberculosis H37Rv | Rv1271c | - | 3e-09 | 34.41% (93) | hypothetical protein Rv1271c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_4150 | - | 1e-12 | 39.78% (93) | hypothetical protein MMAR_4150 |
| M. avium 104 | MAV_3988 | - | 2e-13 | 37.00% (100) | hypothetical protein MAV_3988 |
| M. smegmatis MC2 155 | MSMEG_4689 | - | 6e-06 | 32.08% (106) | hypothetical protein MSMEG_4689 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_4013 | - | 7e-13 | 39.78% (93) | hypothetical protein MUL_4013 |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_1127|M.vanbaalenii_PYR-1 MPDRHSVLRARTAVDVFAEVRFGLQRLDPPGIDMTTLTRLSAIAAMAAAA
Mflv_5185|M.gilvum_PYR-GCK -----------------------------------MFARRAASAGIAAVA
Mb1302c|M.bovis_AF2122/97 --------------------------MLSPLSPRIIAAFTTAVGAAAIGL
Rv1271c|M.tuberculosis_H37Rv --------------------------MLSPLSPRIIAAFTTAVGAAAIGL
MMAR_4150|M.marinum_M --------------------------MFS---TRITAAVTTAIGAAAVGL
MUL_4013|M.ulcerans_Agy99 --------------------------MFS---TRITAAVTTAIGAAAVGL
MAV_3988|M.avium_104 ------------MYHTRAGGRDASAARLYEDEAMRMLALLAGFAALAGMA
MSMEG_4689|M.smegmatis_MC2_155 -------------------------MSARLPAVLVTAAALAAGVGATPAH
: :. . :
Mvan_1127|M.vanbaalenii_PYR-1 AVALAASAAAAPQDAVFLDRLQKVGITSSNPYATVHGGYDVCRQLDL-GT
Mflv_5185|M.gilvum_PYR-GCK AVGLAAPASASPEVAVFLDRLQKVGITSSNPYATIYDAYAVCRELDR-GT
Mb1302c|M.bovis_AF2122/97 AVATAGTAGANTKDEAFIAQMESIGVTFSSPQVATQQAQLVCKKLAS-GE
Rv1271c|M.tuberculosis_H37Rv AVATAGTAGANTKDEAFIAQMESIGVTFSSPQVATQQAQLVCKKLAS-GE
MMAR_4150|M.marinum_M AIATAGSAAASTDDTAFISQMESVGVTFSSPQAADREGHQVCQELAS-GK
MUL_4013|M.ulcerans_Agy99 AIATAGSAAASTDDTAFISQMESVGVTFSSPQAADREGHQVCQELAS-GK
MAV_3988|M.avium_104 APAHADPAGTTGSDASFLAALNQAGITYQNPATAIEVGKRACELMDQ-GS
MSMEG_4689|M.smegmatis_MC2_155 ADPAAPGAPGNPVDRSFLTALNNAGVGYADPDAAVRLGQDVCPMLVEPGK
* * * *: ::. *: .* .: . .* : *
Mvan_1127|M.vanbaalenii_PYR-1 PHSQIVGFVLSDNPDLDWDSAADYVVLANMTYCPPV--------------
Mflv_5185|M.gilvum_PYR-GCK SPTQVVGFVLGDNPDLDWEAAADYVVLANMTYCPPV--------------
Mb1302c|M.bovis_AF2122/97 TGTEIAEEVLS-QTNLTTKQAAYFVVDATKAYCPQYASQLT---------
Rv1271c|M.tuberculosis_H37Rv TGTEIAEEVLS-QTNLTTKQAAYFVVDATKAYCPQYASQLT---------
MMAR_4150|M.marinum_M TGTDIAEEILS-QTDLTSKQAAYFVVYATKDYCPQYASQLA---------
MUL_4013|M.ulcerans_Agy99 TGTDIAEEILS-QTDLTSKQAAYFVVYATKDYCPQYASQLA---------
MAV_3988|M.avium_104 PQVEVIKNVSSSNPGFTVDGAAQFTMIAASAYCPQHLGQRVGPG------
MSMEG_4689|M.smegmatis_MC2_155 NFASVVTKLRGDRSVVSPDVAAFFAGIAISMYCPQMISGIGDGTLLRMLA
.: : . .. . . ** :. * ***
Mvan_1127|M.vanbaalenii_PYR-1 ---------
Mflv_5185|M.gilvum_PYR-GCK ---------
Mb1302c|M.bovis_AF2122/97 ---------
Rv1271c|M.tuberculosis_H37Rv ---------
MMAR_4150|M.marinum_M ---------
MUL_4013|M.ulcerans_Agy99 ---------
MAV_3988|M.avium_104 ---------
MSMEG_4689|M.smegmatis_MC2_155 VPGLGALGR