For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
YHTRAGGRDASAARLYEDEAMRMLALLAGFAALAGMAAPAHADPAGTTGSDASFLAALNQAGITYQNPAT AIEVGKRACELMDQGSPQVEVIKNVSSSNPGFTVDGAAQFTMIAASAYCPQHLGQRVGPG*
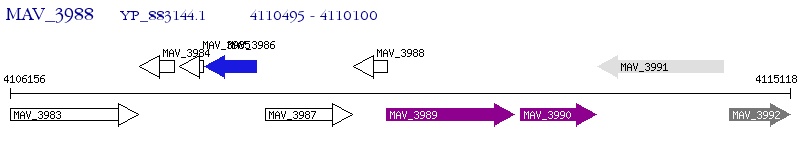
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_3988 | - | - | 100% (131) | hypothetical protein MAV_3988 |
| M. avium 104 | MAV_2912 | - | 6e-24 | 51.46% (103) | hypothetical protein MAV_2912 |
| M. avium 104 | MAV_2904 | - | 1e-22 | 47.17% (106) | hypothetical protein MAV_2904 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1840 | - | 7e-23 | 50.96% (104) | hypothetical protein Mb1840 |
| M. gilvum PYR-GCK | Mflv_5185 | - | 3e-10 | 33.67% (98) | hypothetical protein Mflv_5185 |
| M. tuberculosis H37Rv | Rv1810 | - | 7e-23 | 50.96% (104) | hypothetical protein Rv1810 |
| M. leprae Br4923 | MLBr_00676 | - | 3e-07 | 30.93% (97) | hypothetical protein MLBr_00676 |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_2686 | - | 1e-22 | 46.60% (103) | hypothetical protein MMAR_2686 |
| M. smegmatis MC2 155 | MSMEG_4689 | - | 1e-12 | 42.06% (107) | hypothetical protein MSMEG_4689 |
| M. thermoresistible (build 8) | TH_1573 | - | 4e-05 | 31.43% (105) | conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_3300 | - | 8e-22 | 49.00% (100) | hypothetical protein MUL_3300 |
| M. vanbaalenii PYR-1 | Mvan_1127 | - | 3e-13 | 37.00% (100) | hypothetical protein Mvan_1127 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_5185|M.gilvum_PYR-GCK -----------------------------------MFARRAASAGIAAVA
Mvan_1127|M.vanbaalenii_PYR-1 MPDRHSVLRARTAVDVFAEVRFGLQRLDPPGIDMTTLTRLSAIAAMAAAA
MAV_3988|M.avium_104 ----------------MYHTRAGGRDASAARLYEDEAMRMLALLAG-FAA
MUL_3300|M.ulcerans_Agy99 -------------------------------------MKLLVGVTG-LAV
Mb1840|M.bovis_AF2122/97 ----------------MQLQRTMGQCR---------PMRMLVALLLSAAT
Rv1810|M.tuberculosis_H37Rv ----------------MQLQRTMGQCR---------PMRMLVALLLSAAT
MMAR_2686|M.marinum_M -------------------------------------MKTLFALLG-LSA
MSMEG_4689|M.smegmatis_MC2_155 -----------------------------------MSARLPAVLVTAAAL
MLBr_00676|M.leprae_Br4923 ----MFLRLMLSALKALQRLGAVMNSLARIDHWIWLFRCQPLTIRLLVAT
TH_1573|M.thermoresistible__bu ------------------------------------MRGYRVFAAALVLS
Mflv_5185|M.gilvum_PYR-GCK AVGL-AAPASASPE---------VAVFLDRLQKVGITSSNPYATIYDAYA
Mvan_1127|M.vanbaalenii_PYR-1 AVAL-AASAAAAPQ---------DAVFLDRLQKVGITSSNPYATVHGGYD
MAV_3988|M.avium_104 LAGM-AAPAHADPAGT----TGSDASFLAALNQAGITYQNPATAIEVGKR
MUL_3300|M.ulcerans_Agy99 MVGL-AAPAHADAN---------DDAFLVALGKAGITYPDAARAIAAAKW
Mb1840|M.bovis_AF2122/97 MIGL-AAPGKADPT-------GDDAAFLAALDQAGITYADPGHAITAAKA
Rv1810|M.tuberculosis_H37Rv MIGL-AAPGKADPT-------GDDAAFLAALDQAGITYADPGHAITAAKA
MMAR_2686|M.marinum_M MIWL-AAPAHAHPDEG----GGDDVGFLAALQNAGITYSNPDQAIGSAKA
MSMEG_4689|M.smegmatis_MC2_155 AAGVGATPAHADPAAPGAPGNPVDRSFLTALNNAGVGYADPDAAVRLGQD
MLBr_00676|M.leprae_Br4923 AALFTAATAFEVPAEAD----AIDDTFIKALNHAGVNFGEPRSAMTMGHY
TH_1573|M.thermoresistible__bu ASALSASVSMAGPAH------ADQYDFLIELDNAGVWYPSSVDMVDIGKE
. *: . . *: * :.*: .. : .
Mflv_5185|M.gilvum_PYR-GCK VCRELDR-GTSPTQVVGFVLGDNPDLDWEAAADYVVLANMTYCPPV----
Mvan_1127|M.vanbaalenii_PYR-1 VCRQLDL-GTPHSQIVGFVLSDNPDLDWDSAADYVVLANMTYCPPV----
MAV_3988|M.avium_104 ACELMDQ-GSPQVEVIKNVSSSNPGFTVDGAAQFTMIAASAYCPQHLGQR
MUL_3300|M.ulcerans_Agy99 ICQQVNN-GTQTVDVVKQVQSSNSGLQEDGAAKFTAIAANAYCPAILSGH
Mb1840|M.bovis_AF2122/97 MCGLCAN-GVTGLQLVADLRDYNPGLTMDSAAKFAAIASGAYCPEHLEHH
Rv1810|M.tuberculosis_H37Rv MCGLCAN-GVTGLQLVADLRDYNPGLTMDSAAKFAAIASGAYCPEHLEHH
MMAR_2686|M.marinum_M VCWCLDG-GESGLELVHDVKTHNPGFNMEAASQFAVLAATYYCPHHLSHA
MSMEG_4689|M.smegmatis_MC2_155 VCPMLVEPGKNFASVVTKLRGDRSVVSPDVAAFFAGIAISMYCPQMISGI
MLBr_00676|M.leprae_Br4923 VCPILAKSGGNFAAAVQRIRG-NSDMSPQMAETFAKIAISIYCPTMMANV
TH_1573|M.thermoresistible__bu VCHELRH-RVSPGLVLDKLVN--TGFAPAEAVIVLASAVTHMCVDVQPAV
* : : . . * * *
Mflv_5185|M.gilvum_PYR-GCK -------------------
Mvan_1127|M.vanbaalenii_PYR-1 -------------------
MAV_3988|M.avium_104 VG--PG-------------
MUL_3300|M.ulcerans_Agy99 GANPPG-------------
Mb1840|M.bovis_AF2122/97 PS-----------------
Rv1810|M.tuberculosis_H37Rv PS-----------------
MMAR_2686|M.marinum_M -------------------
MSMEG_4689|M.smegmatis_MC2_155 GDGTLLRMLAVPGLGALGR
MLBr_00676|M.leprae_Br4923 ASGNLPSLPPGPGIPGI--
TH_1573|M.thermoresistible__bu VSWVNSQQYSAPV------