For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MFSTRITAAVTTAIGAAAVGLAIATAGSAAASTDDTAFISQMESVGVTFSSPQAADREGHQVCQELASGK TGTDIAEEILSQTDLTSKQAAYFVVYATKDYCPQYASQLA
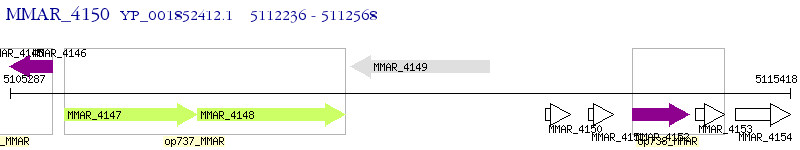
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_4150 | - | - | 100% (110) | hypothetical protein MMAR_4150 |
| M. marinum M | MMAR_4151 | - | 3e-44 | 80.73% (109) | hypothetical protein MMAR_4151 |
| M. marinum M | MMAR_1233 | - | 2e-15 | 38.89% (108) | hypothetical protein MMAR_1233 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1302c | - | 3e-40 | 72.90% (107) | hypothetical protein Mb1302c |
| M. gilvum PYR-GCK | Mflv_2012 | - | 3e-11 | 39.08% (87) | hypothetical protein Mflv_2012 |
| M. tuberculosis H37Rv | Rv1271c | - | 3e-40 | 72.90% (107) | hypothetical protein Rv1271c |
| M. leprae Br4923 | MLBr_00676 | - | 1e-07 | 32.65% (98) | hypothetical protein MLBr_00676 |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. avium 104 | MAV_1419 | - | 5e-25 | 53.15% (111) | hypothetical protein MAV_1419 |
| M. smegmatis MC2 155 | MSMEG_5766 | - | 3e-09 | 32.41% (108) | hypothetical protein MSMEG_5766 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_4013 | - | 5e-57 | 100.00% (110) | hypothetical protein MUL_4013 |
| M. vanbaalenii PYR-1 | Mvan_1127 | - | 6e-13 | 39.78% (93) | hypothetical protein Mvan_1127 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4150|M.marinum_M ---------------------------MFS---TRITAAVTTAIGAAAVG
MUL_4013|M.ulcerans_Agy99 ---------------------------MFS---TRITAAVTTAIGAAAVG
Mb1302c|M.bovis_AF2122/97 ---------------------------MLSPLSPRIIAAFTTAVGAAAIG
Rv1271c|M.tuberculosis_H37Rv ---------------------------MLSPLSPRIIAAFTTAVGAAAIG
MAV_1419|M.avium_104 ---------------------------MFSR--RIITVLITAAIGLAAVG
Mvan_1127|M.vanbaalenii_PYR-1 -MPDRHSVLRARTAVDVFAEVRFGLQRLDPPGIDMTTLTRLSAIAAMAAA
MSMEG_5766|M.smegmatis_MC2_155 ---------------------------------MIFRRQTAWAVAAAAIA
Mflv_2012|M.gilvum_PYR-GCK ----------------------------------MRGTFFITTLAAAGLA
MLBr_00676|M.leprae_Br4923 MFLRLMLSALKALQRLGAVMNSLARIDHWIWLFRCQPLTIRLLVATAALF
:. .
MMAR_4150|M.marinum_M LAIAT-AGSAAASTDDTAFISQMESVGVTFSSPQAADREGHQVCQELASG
MUL_4013|M.ulcerans_Agy99 LAIAT-AGSAAASTDDTAFISQMESVGVTFSSPQAADREGHQVCQELASG
Mb1302c|M.bovis_AF2122/97 LAVAT-AGTAGANTKDEAFIAQMESIGVTFSSPQVATQQAQLVCKKLASG
Rv1271c|M.tuberculosis_H37Rv LAVAT-AGTAGANTKDEAFIAQMESIGVTFSSPQVATQQAQLVCKKLASG
MAV_1419|M.avium_104 LAGPA-AART-VQTGDEMFLAQMRSLGVSFPSPQEAVREGHQVCAELSAG
Mvan_1127|M.vanbaalenii_PYR-1 AAVAL-AASAAAAPQDAVFLDRLQKVGITSSNPYATVHGGYDVCRQLDLG
MSMEG_5766|M.smegmatis_MC2_155 SGVAFGTAPAHASVEDDRYLKIVEQLDIP-GEPEAAIQVGHEICKAVEAG
Mflv_2012|M.gilvum_PYR-GCK TALAA---PAFADETDDIFIAALEQEGIPFSTAGNAIELAGAVCEYAAAG
MLBr_00676|M.leprae_Br4923 TAATAFEVPAEADAIDDTFIKALNHAGVNFGEPRSAMTMGHYVCPILAKS
. . : . * :: :. .: . : . :* .
MMAR_4150|M.marinum_M KTG-----TDIAEEILS-QTDLTSKQAAYFVVYATKDYCPQYASQLA---
MUL_4013|M.ulcerans_Agy99 KTG-----TDIAEEILS-QTDLTSKQAAYFVVYATKDYCPQYASQLA---
Mb1302c|M.bovis_AF2122/97 ETG-----TEIAEEVLS-QTNLTTKQAAYFVVDATKAYCPQYASQLT---
Rv1271c|M.tuberculosis_H37Rv ETG-----TEIAEEVLS-QTNLTTKQAAYFVVDATKAYCPQYASQLT---
MAV_1419|M.avium_104 KTP-----TAVTVAVFR-QTNLTPPQAAGLVTAATQAYCPQFSGQPA---
Mvan_1127|M.vanbaalenii_PYR-1 TPH-----SQIVGFVLSDNPDLDWDSAADYVVLANMTYCPPV--------
MSMEG_5766|M.smegmatis_MC2_155 KIEPARTVRGVINHLMN-QAGISKGQAAGLVRGAVSVYCPQYTSLVGR--
Mflv_2012|M.gilvum_PYR-GCK QDK-----TQIALEIME-PAGWSPEQSGFFVGAATQSYCP----------
MLBr_00676|M.leprae_Br4923 GGN-----FAAAVQRIRGNSDMSPQMAETFAKIAISIYCPTMMANVASGN
: .. : . * ***
MMAR_4150|M.marinum_M -------------
MUL_4013|M.ulcerans_Agy99 -------------
Mb1302c|M.bovis_AF2122/97 -------------
Rv1271c|M.tuberculosis_H37Rv -------------
MAV_1419|M.avium_104 -------------
Mvan_1127|M.vanbaalenii_PYR-1 -------------
MSMEG_5766|M.smegmatis_MC2_155 -------------
Mflv_2012|M.gilvum_PYR-GCK -------------
MLBr_00676|M.leprae_Br4923 LPSLPPGPGIPGI