For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PESTEKEQLSIGILAGSRKPDERRLPIHPAHLGRIDRAVASRVFLEHGYGSRFGATDAGLSDHVGGIRTR EELFAECDVILLPKVQAEDLAECSVGQIVWGWPHCVQDADLTQVAIDRKLTLVAFEAMNHWRPDGGFSLH VFHKNNELAGYCSVLHSTQLAGVTGSYGRRLSAAILGFGATARGAVTALNALGVDDVHVLTNRDTAAVGS PIHSTQISQMGRDEGAPDRIWADVPDGRVPLAQFLADRDIVVNCVLQDPNAPLIFATEDDLSSFAPRSLI VDVSCDVGMGFSWARPTSFARPVVTLPNDVLYYAVDHSPSYLWDSATWEISEALLPHLRPLLEGPATWAV TPTIARAIEIHDGVVLNEDILSYQHRDSAYPHEVGTGHR*
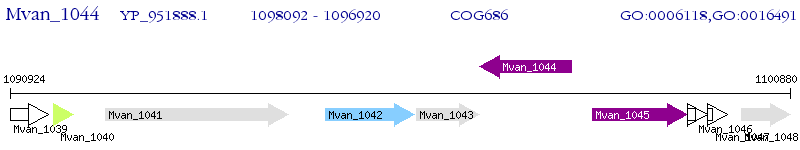
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_1044 | - | - | 100% (390) | NAD(P) transhydrogenase subunit alpha |
| M. vanbaalenii PYR-1 | Mvan_2355 | - | 2e-13 | 21.93% (374) | alanine dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2803 | aldb | 1e-08 | 24.19% (124) | L-alanine dehydrogenase |
| M. gilvum PYR-GCK | Mflv_5223 | - | 0.0 | 83.33% (390) | NAD(P) transhydrogenase subunit alpha |
| M. tuberculosis H37Rv | Rv2780 | ald | 5e-14 | 22.85% (372) | secreted L-alanine dehydrogenase ALD (40 kDa antigen) (TB43) |
| M. leprae Br4923 | MLBr_01532 | ald | 1e-10 | 22.31% (372) | L-alanine dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3100 | - | 6e-11 | 23.16% (367) | L-alanine dehydrogenase (ALD) |
| M. marinum M | MMAR_1928 | ald | 9e-09 | 20.11% (373) | secreted L-alanine dehydrogenase Ald (40 kDa antigen) |
| M. avium 104 | MAV_3674 | ald | 5e-08 | 21.84% (380) | alanine dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_2659 | ald | 2e-10 | 21.82% (362) | alanine dehydrogenase |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_2154 | ald | 8e-09 | 20.11% (373) | secreted L-alanine dehydrogenase Ald (40 kDa antigen) |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2803|M.bovis_AF2122/97 --------------------------------------------------
Rv2780|M.tuberculosis_H37Rv ---------MRVGIPTETKN-NEFRVAITPAGVAELTRR-GHEVLIQAGA
MLBr_01532|M.leprae_Br4923 ---------MRVGIPTETKN-NEFRVAITPAGVAALTHR-GHEVLVQAGA
MAV_3674|M.avium_104 ---------MRVGVPTETKT-NEFRVALTPAGVAELTRR-GHEVLVQSGA
MMAR_1928|M.marinum_M ---------MRVGIPTETKD-NEFRVAITPAGVAALTKR-GHEVLIQAGA
MUL_2154|M.ulcerans_Agy99 ---------MRVGIPTETKD-NEFRVAITPAGVAALTKR-GHEVLIQVGA
MSMEG_2659|M.smegmatis_MC2_155 ---------MLVGIPTEIKN-NEYRVAITPAGVAELTRR-GHEVIIQAGA
MAB_3100|M.abscessus_ATCC_1997 ---------MRVGIPTEIKN-NEYRVATTPAGVAELTRR-GHEVIIQSGA
Mvan_1044|M.vanbaalenii_PYR-1 MPESTEKEQLSIGILAGSRKPDERRLPIHPAHLGRIDRAVASRVFLEHGY
Mflv_5223|M.gilvum_PYR-GCK MSEKAEPEQLSIGILATSRKADERRLPIHPAHLSRIGRAVGRRVFLEQGY
Mb2803|M.bovis_AF2122/97 --------------------------------------------------
Rv2780|M.tuberculosis_H37Rv GEGSAITDADFKAAGAQLVGTADQVWADADLLLKVKEPIAAEYGRLRHGQ
MLBr_01532|M.leprae_Br4923 GEGSAIADAEFKAAGAQLISTPEQVWTEVDLLLKVKEPMPAEYVHLRHGQ
MAV_3674|M.avium_104 GEGSSIPDSEFKAAGAQLADTAEQVWADAELLLKVKEPVPAEYDLLRRNQ
MMAR_1928|M.marinum_M GEGSAISDSDFKAAGAQLISTADQVWADADLLLKVKEPIESEYGRLRRGQ
MUL_2154|M.ulcerans_Agy99 GEGSAISDSDFKAAGAQLISTADQVWADADLLLKVKEPIESEYGRLRRGQ
MSMEG_2659|M.smegmatis_MC2_155 GEGSAISDRDFKAAGAEIVNTADQVWSEAELLLKVKEPIEPEYSRMRKGQ
MAB_3100|M.abscessus_ATCC_1997 GDGSSISDVDFKAAGAQVINGAEQVWAEADLLLKVKEPIEPEYALMRRGQ
Mvan_1044|M.vanbaalenii_PYR-1 GSRFGATDAGLSDHVGGIR-TREELFAECDVILLP-KVQAEDLAECSVGQ
Mflv_5223|M.gilvum_PYR-GCK GSHFGATDDAVSDHVGGVR-SRKELIAECDVILLP-KVHAEDLAECRTGQ
Mb2803|M.bovis_AF2122/97 -----------------------------------------------MSE
Rv2780|M.tuberculosis_H37Rv ILFTFLHLAASRACTDALLDSGTTSIAYETVQTADG----ALPLLAPMSE
MLBr_01532|M.leprae_Br4923 ILFTYLHLAASRPCTDALLASGTTSIAYETVQTADG----ALPLLAPMSE
MAV_3674|M.avium_104 ILFTFLHLAASRACTEALLSSGTTSIAYETVQTADG----TLPLLAPMSE
MMAR_1928|M.marinum_M TLFTYLHLAASRPCTDALLKSGTTSIAYETVQTADG----ALPLLAPMSE
MUL_2154|M.ulcerans_Agy99 TLFTYLHLAASRPCTDALLKSGTTSIAYETVQTADG----ALPLLAPMSE
MSMEG_2659|M.smegmatis_MC2_155 TLFTYLHLAASKPCTDALLASGTTSIAYETVQTAEG----ALPLLAPMSE
MAB_3100|M.abscessus_ATCC_1997 TLFTYLHLAASLPCTQALLKSETTSIAYETVQTSDG----ALPLLAPMSE
Mvan_1044|M.vanbaalenii_PYR-1 IVWGWPHCVQDADLTQVAIDRKLTLVAFEAMNHWRPDGGFSLHVFHKNNE
Mflv_5223|M.gilvum_PYR-GCK IIWGWPHCVQDTGLTQVAIDRKLTLIAFEAMNHWRADGGFGLHVFHKNNE
.*
Mb2803|M.bovis_AF2122/97 VAGRLAAQVGAYHLMRTQGGRG-VLMGGVPGVEP-ADVVVIGAGTAGYNA
Rv2780|M.tuberculosis_H37Rv VAGRLAAQVGAYHLMRTQGGRG-VLMGGVPGVEP-ADVVVIGAGTAGYNA
MLBr_01532|M.leprae_Br4923 VAGRLSAQVGAYHLMRTHRGCG-VLMGGVPGVEP-ADVVVIGGGTAGYNA
MAV_3674|M.avium_104 VAGRLSAQVGAYHLMRTHGGRG-VLMGGVPGVKP-ADVVVVGAGTAGYNA
MMAR_1928|M.marinum_M VAGRLSAQAGAYHLMRTHGGRG-VLMGGVPGVKP-ADVVVIGAGTAGYNA
MUL_2154|M.ulcerans_Agy99 VAGRLSAQAGAYHLMRTHGGRG-VLMGGVPGVKP-ADVVVIGAGTAGYNA
MSMEG_2659|M.smegmatis_MC2_155 VAGRLSAQVGAYHLMRSYGGRG-VLMGGVPGVAP-AEVVVIGAGTAGYNA
MAB_3100|M.abscessus_ATCC_1997 VAGRLAAQVGAYHLMTPVGGRG-LVMGGVPGVGP-ADVVVIGGGVAGYNA
Mvan_1044|M.vanbaalenii_PYR-1 LAG-YCSVLHSTQLAGVTGSYGRRLSAAILGFGATARGAVTALNALGVDD
Mflv_5223|M.gilvum_PYR-GCK LAG-YCSVLHATQLAGVTGTYGRRLSAAVLGFGATARGAVTALNALGVDD
:** .: : :* * : ..: *. . * .* . .. * :
Mb2803|M.bovis_AF2122/97 ARIANGMGATVTVLDINIDKLRQLDAEFCGRIHTRYSS---AYELEGAVK
Rv2780|M.tuberculosis_H37Rv ARIANGMGATVTVLDINIDKLRQLDAEFCGRIHTRYSS---AYELEGAVK
MLBr_01532|M.leprae_Br4923 ARIANGMGATVTVFDVNINRLRELDAEFGGRLRTRYSS---AYDLEGAVK
MAV_3674|M.avium_104 ARVAGGMGAMVTVLDVNINKLRLLDAEFDGQVRTRYSS---AYDLEGTVK
MMAR_1928|M.marinum_M ARVANGMGAMVTVLDVNINKLRQIDAEFGGRVRTRYSS---TLDLEDAAV
MUL_2154|M.ulcerans_Agy99 ARVANGMGAMVTVLDVNINKLRQIDAEFGGRVRTRYSS---TLDLEDAAV
MSMEG_2659|M.smegmatis_MC2_155 ARVAAGMGAHVTVFDLNINTLRRVDGEFGGRIETRYSS---SLELEEAVK
MAB_3100|M.abscessus_ATCC_1997 ARIAAGMGAHVTVFDVNLGKLRELDGEFGGTIRTRYSS---TMELEGAVK
Mvan_1044|M.vanbaalenii_PYR-1 VHVLTNRDTAAVGSPIHSTQISQMGRDEGAPDRIWADVPDGRVPLAQFLA
Mflv_5223|M.gilvum_PYR-GCK VHVLTNRDTAAVGSPIHSTQISQMGPEPDAPDRIWADTPDGRIPVAQFLS
.:: . .: .. :: : :. : . . . :
Mb2803|M.bovis_AF2122/97 RADLVIGAVLVPGAKAPKLVSNSLVAHMKPGAVLVDIAIDQGGCFEGSRP
Rv2780|M.tuberculosis_H37Rv RADLVIGAVLVPGAKAPKLVSNSLVAHMKPGAVLVDIAIDQGGCFEGSRP
MLBr_01532|M.leprae_Br4923 HADLVIGAVLVPGAKAPRLISNSLVAHMKPGAVLVDIAIDQGGCFEDSQP
MAV_3674|M.avium_104 RADLVIGAVLLPGAKAPNLISNSLVAQMKSGAVLVDISIDQGGCFEDSRP
MMAR_1928|M.marinum_M NADLLIGAVLVPGAKAPKLIPNSLVARMKKGAVLVDISIDQGGCFEDSRP
MUL_2154|M.ulcerans_Agy99 NADLLIGAVLVPGAKAPKLIPNSLVARMKKGAVLVDISIDQGGCFEDSRP
MSMEG_2659|M.smegmatis_MC2_155 KADLVIGAVLVPGAKAPKLVTNSTVAHMKPGAVLVDIAIDQGGCFEDSRP
MAB_3100|M.abscessus_ATCC_1997 RADLVIGAVLIPGAKAPKLISNSLVSQMKPGSVLVDIAIDQGGCFADSRP
Mvan_1044|M.vanbaalenii_PYR-1 DRDIVVNCVLQDPNAPLIFATEDDLSSFAPRSLIVDVSCDVGMGFSWARP
Mflv_5223|M.gilvum_PYR-GCK DKDIVVNCVLQDPNAPLTFVSEEDLSSFAPRSLIIDVSCDVGMGFSWARP
*:::..** . : .:. :: : ::::*:: * * * ::*
Mb2803|M.bovis_AF2122/97 TTYDHPTFAVH-DTLFYCVANMPASVPKTSTYALTNATMPYVLELADHGW
Rv2780|M.tuberculosis_H37Rv TTYDHPTFAVH-DTLFYCVANMPASVPKTSTYALTNATMPYVLELADHGW
MLBr_01532|M.leprae_Br4923 TTHDQPTFAVH-DTLFYCVANMPSAVPKTSTFALTNATMPYILKLADQGW
MAV_3674|M.avium_104 TTHDDPTFAVH-DTVFYCVANMPSAVPKTSTVALTNATMPYVLALADRGW
MMAR_1928|M.marinum_M TTHTSPTFTVH-DTVFYCVANMPGAAPRTSTYALTNATMPYVLKLADLGW
MUL_2154|M.ulcerans_Agy99 TTHTSPTFTVH-DTVFYCVANMPGAAPRTSTYALTNATMPYVLKLADLGW
MSMEG_2659|M.smegmatis_MC2_155 TTHDEPTFKVH-DTIFYCVANMPGAVPRTSTFALTNSTMPYVLKLADKGW
MAB_3100|M.abscessus_ATCC_1997 TTHDEPTYTVH-DSVFYCVANMPGAVPRTSTFALTNATMPYVLALADKGW
Mvan_1044|M.vanbaalenii_PYR-1 TSFARPVVTLPNDVLYYAVDHSPSYLWDSATWEISEALLPHLRPLLEG--
Mflv_5223|M.gilvum_PYR-GCK TSFARPVVALPNGLLYYAVDHSPSYLWDSATWEISEALLPHLAPLLEG--
*:. *. : . ::*.* : *. ::* :::: :*:: * :
Mb2803|M.bovis_AF2122/97 RAACRSNPALAKGLSTHEGALLSERVATDLGVPFTEPASVLA---
Rv2780|M.tuberculosis_H37Rv RAACRSNPALAKGLSTHEGALLSERVATDLGVPFTEPASVLA---
MLBr_01532|M.leprae_Br4923 QEACQSDPALAKGLSTHNGALLSEQVATDLGLAFTEPASVLA---
MAV_3674|M.avium_104 RAACRSDHALAKGLSTHDGTLLSQQVAADLGLPCTDPAGVLQQD-
MMAR_1928|M.marinum_M QAACLSDSALAKGLSTHEGALLADHVAADLDLPFTDPANVLG---
MUL_2154|M.ulcerans_Agy99 QAACLSDSALAKGLSTHEGALLADHVAADLDLPFTDPASVLG---
MSMEG_2659|M.smegmatis_MC2_155 QAACASDSALAKGLSTHDGKLLSEAVAKDLDLPFTDAAQFLA---
MAB_3100|M.abscessus_ATCC_1997 RQACRDDAALAQGLSTHEGALLNTHVAHDLGLAFTDPAEVLG---
Mvan_1044|M.vanbaalenii_PYR-1 PATWAVTPTIARAIEIHDGVVLNEDILSYQHRDSAYPHEVGTGHR
Mflv_5223|M.gilvum_PYR-GCK PATWDTMPTLSRAIEIREGVVVNENILTFQHRASAYPHEVSVTAR
: ::::.:. ::* :: : : . .