For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MRVGIPTETKNNEFRVAITPAGVAELTRRGHEVLIQAGAGEGSAITDADFKAAGAQLVGTADQVWADADL LLKVKEPIAAEYGRLRHGQILFTFLHLAASRACTDALLDSGTTSIAYETVQTADGALPLLAPMSEVAGRL AAQVGAYHLMRTQGGRGVLMGGVPGVEPADVVVIGAGTAGYNAARIANGMGATVTVLDINIDKLRQLDAE FCGRIHTRYSSAYELEGAVKRADLVIGAVLVPGAKAPKLVSNSLVAHMKPGAVLVDIAIDQGGCFEGSRP TTYDHPTFAVHDTLFYCVANMPASVPKTSTYALTNATMPYVLELADHGWRAACRSNPALAKGLSTHEGAL LSERVATDLGVPFTEPASVLA
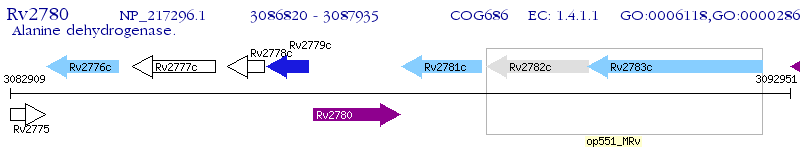
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv2780 | ald | - | 100% (371) | SECRETED L-ALANINE DEHYDROGENASE ALD (40 KDA ANTIGEN) (TB43) |
| M. tuberculosis H37Rv | Rv0155 | pntAa | 2e-34 | 33.44% (326) | NAD(P) transhydrogenase subunit alpha |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2803 | aldb | 1e-136 | 100.00% (239) | L-alanine dehydrogenase |
| M. gilvum PYR-GCK | Mflv_4017 | - | 1e-168 | 78.44% (371) | alanine dehydrogenase |
| M. leprae Br4923 | MLBr_01532 | ald | 0.0 | 85.44% (371) | L-alanine dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3100 | - | 1e-165 | 77.84% (370) | L-alanine dehydrogenase (ALD) |
| M. marinum M | MMAR_1928 | ald | 1e-177 | 82.97% (370) | secreted L-alanine dehydrogenase Ald (40 kDa antigen) |
| M. avium 104 | MAV_3674 | ald | 1e-177 | 82.70% (370) | alanine dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_2659 | ald | 1e-171 | 80.59% (371) | alanine dehydrogenase |
| M. thermoresistible (build 8) | TH_1286 | ald | 9e-47 | 66.94% (121) | SECRETED L-ALANINE DEHYDROGENASE ALD (40 KDA ANTIGEN) (TB43) |
| M. ulcerans Agy99 | MUL_2154 | ald | 1e-177 | 82.97% (370) | secreted L-alanine dehydrogenase Ald (40 kDa antigen) |
| M. vanbaalenii PYR-1 | Mvan_2355 | - | 1e-167 | 78.71% (371) | alanine dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4017|M.gilvum_PYR-GCK MRVGIPTEIKNNEFRVAITPSGVAELVQRGHEVLIQSGAGDGSSISDADF
Mvan_2355|M.vanbaalenii_PYR-1 MRVGIPTEIKNNEYRVAVTPAGVAELVRRGHEVLIQSGAGDGSAISDADF
Rv2780|M.tuberculosis_H37Rv MRVGIPTETKNNEFRVAITPAGVAELTRRGHEVLIQAGAGEGSAITDADF
Mb2803|M.bovis_AF2122/97 --------------------------------------------------
MLBr_01532|M.leprae_Br4923 MRVGIPTETKNNEFRVAITPAGVAALTHRGHEVLVQAGAGEGSAIADAEF
MAV_3674|M.avium_104 MRVGVPTETKTNEFRVALTPAGVAELTRRGHEVLVQSGAGEGSSIPDSEF
MMAR_1928|M.marinum_M MRVGIPTETKDNEFRVAITPAGVAALTKRGHEVLIQAGAGEGSAISDSDF
MUL_2154|M.ulcerans_Agy99 MRVGIPTETKDNEFRVAITPAGVAALTKRGHEVLIQVGAGEGSAISDSDF
MAB_3100|M.abscessus_ATCC_1997 MRVGIPTEIKNNEYRVATTPAGVAELTRRGHEVIIQSGAGDGSSISDVDF
MSMEG_2659|M.smegmatis_MC2_155 MLVGIPTEIKNNEYRVAITPAGVAELTRRGHEVIIQAGAGEGSAISDRDF
TH_1286|M.thermoresistible__bu --------------------------------------------------
Mflv_4017|M.gilvum_PYR-GCK KRAGAQLITGADEVWSEADLLLKVKEPIEAEYSKMREGQTLFTYLHLAAS
Mvan_2355|M.vanbaalenii_PYR-1 KRAGGQLVTGADEVWAEAELLLKVKEPIEAEYSRMREGQTLFTYLHLAAS
Rv2780|M.tuberculosis_H37Rv KAAGAQLVGTADQVWADADLLLKVKEPIAAEYGRLRHGQILFTFLHLAAS
Mb2803|M.bovis_AF2122/97 --------------------------------------------------
MLBr_01532|M.leprae_Br4923 KAAGAQLISTPEQVWTEVDLLLKVKEPMPAEYVHLRHGQILFTYLHLAAS
MAV_3674|M.avium_104 KAAGAQLADTAEQVWADAELLLKVKEPVPAEYDLLRRNQILFTFLHLAAS
MMAR_1928|M.marinum_M KAAGAQLISTADQVWADADLLLKVKEPIESEYGRLRRGQTLFTYLHLAAS
MUL_2154|M.ulcerans_Agy99 KAAGAQLISTADQVWADADLLLKVKEPIESEYGRLRRGQTLFTYLHLAAS
MAB_3100|M.abscessus_ATCC_1997 KAAGAQVINGAEQVWAEADLLLKVKEPIEPEYALMRRGQTLFTYLHLAAS
MSMEG_2659|M.smegmatis_MC2_155 KAAGAEIVNTADQVWSEAELLLKVKEPIEPEYSRMRKGQTLFTYLHLAAS
TH_1286|M.thermoresistible__bu --------------------------------------------------
Mflv_4017|M.gilvum_PYR-GCK KPCTDALMASGTTSIAYETVQLTDGSLPLLAPMSEVAGRLSAQVGSYHLM
Mvan_2355|M.vanbaalenii_PYR-1 RSCTDALMASRTTSIAYETVQTADGALPLLAPMSEVAGRLSAQVGAYHLM
Rv2780|M.tuberculosis_H37Rv RACTDALLDSGTTSIAYETVQTADGALPLLAPMSEVAGRLAAQVGAYHLM
Mb2803|M.bovis_AF2122/97 --------------------------------MSEVAGRLAAQVGAYHLM
MLBr_01532|M.leprae_Br4923 RPCTDALLASGTTSIAYETVQTADGALPLLAPMSEVAGRLSAQVGAYHLM
MAV_3674|M.avium_104 RACTEALLSSGTTSIAYETVQTADGTLPLLAPMSEVAGRLSAQVGAYHLM
MMAR_1928|M.marinum_M RPCTDALLKSGTTSIAYETVQTADGALPLLAPMSEVAGRLSAQAGAYHLM
MUL_2154|M.ulcerans_Agy99 RPCTDALLKSGTTSIAYETVQTADGALPLLAPMSEVAGRLSAQAGAYHLM
MAB_3100|M.abscessus_ATCC_1997 LPCTQALLKSETTSIAYETVQTSDGALPLLAPMSEVAGRLAAQVGAYHLM
MSMEG_2659|M.smegmatis_MC2_155 KPCTDALLASGTTSIAYETVQTAEGALPLLAPMSEVAGRLSAQVGAYHLM
TH_1286|M.thermoresistible__bu --------------------------------------------------
Mflv_4017|M.gilvum_PYR-GCK RSHGGRGVLMGGVPGVAPAEVVVIGGGVAGYNAARIAKGMGAHVNVFDVN
Mvan_2355|M.vanbaalenii_PYR-1 RTQGGRGVLMGGVPGVAPAKVVVIGGGMAGDNAAAVAWGMGAHVTVFDLN
Rv2780|M.tuberculosis_H37Rv RTQGGRGVLMGGVPGVEPADVVVIGAGTAGYNAARIANGMGATVTVLDIN
Mb2803|M.bovis_AF2122/97 RTQGGRGVLMGGVPGVEPADVVVIGAGTAGYNAARIANGMGATVTVLDIN
MLBr_01532|M.leprae_Br4923 RTHRGCGVLMGGVPGVEPADVVVIGGGTAGYNAARIANGMGATVTVFDVN
MAV_3674|M.avium_104 RTHGGRGVLMGGVPGVKPADVVVVGAGTAGYNAARVAGGMGAMVTVLDVN
MMAR_1928|M.marinum_M RTHGGRGVLMGGVPGVKPADVVVIGAGTAGYNAARVANGMGAMVTVLDVN
MUL_2154|M.ulcerans_Agy99 RTHGGRGVLMGGVPGVKPADVVVIGAGTAGYNAARVANGMGAMVTVLDVN
MAB_3100|M.abscessus_ATCC_1997 TPVGGRGLVMGGVPGVGPADVVVIGGGVAGYNAARIAAGMGAHVTVFDVN
MSMEG_2659|M.smegmatis_MC2_155 RSYGGRGVLMGGVPGVAPAEVVVIGAGTAGYNAARVAAGMGAHVTVFDLN
TH_1286|M.thermoresistible__bu --------------------------------------------------
Mflv_4017|M.gilvum_PYR-GCK INTLRKIDNENNGAIETRYSSSLDLEDAVKHADLVIGAVLVPGARAPKLV
Mvan_2355|M.vanbaalenii_PYR-1 VNTLRKIDAEYGGAIETRYSSRLDLEDAVKQADLVIGAVLVPGHKAPKLV
Rv2780|M.tuberculosis_H37Rv IDKLRQLDAEFCGRIHTRYSSAYELEGAVKRADLVIGAVLVPGAKAPKLV
Mb2803|M.bovis_AF2122/97 IDKLRQLDAEFCGRIHTRYSSAYELEGAVKRADLVIGAVLVPGAKAPKLV
MLBr_01532|M.leprae_Br4923 INRLRELDAEFGGRLRTRYSSAYDLEGAVKHADLVIGAVLVPGAKAPRLI
MAV_3674|M.avium_104 INKLRLLDAEFDGQVRTRYSSAYDLEGTVKRADLVIGAVLLPGAKAPNLI
MMAR_1928|M.marinum_M INKLRQIDAEFGGRVRTRYSSTLDLEDAAVNADLLIGAVLVPGAKAPKLI
MUL_2154|M.ulcerans_Agy99 INKLRQIDAEFGGRVRTRYSSTLDLEDAAVNADLLIGAVLVPGAKAPKLI
MAB_3100|M.abscessus_ATCC_1997 LGKLRELDGEFGGTIRTRYSSTMELEGAVKRADLVIGAVLIPGAKAPKLI
MSMEG_2659|M.smegmatis_MC2_155 INTLRRVDGEFGGRIETRYSSSLELEEAVKKADLVIGAVLVPGAKAPKLV
TH_1286|M.thermoresistible__bu -------------------------------------------------V
:
Mflv_4017|M.gilvum_PYR-GCK SNSTVAAMKPGAVLVDIAIDQGGCFEDSRPTTHDDPTFAVHDTVFYCVAN
Mvan_2355|M.vanbaalenii_PYR-1 TNSTVAAMKPGAVLVDIAIDQGGCFEDSRPTTHDDPTFTVHDTVFYCVAN
Rv2780|M.tuberculosis_H37Rv SNSLVAHMKPGAVLVDIAIDQGGCFEGSRPTTYDHPTFAVHDTLFYCVAN
Mb2803|M.bovis_AF2122/97 SNSLVAHMKPGAVLVDIAIDQGGCFEGSRPTTYDHPTFAVHDTLFYCVAN
MLBr_01532|M.leprae_Br4923 SNSLVAHMKPGAVLVDIAIDQGGCFEDSQPTTHDQPTFAVHDTLFYCVAN
MAV_3674|M.avium_104 SNSLVAQMKSGAVLVDISIDQGGCFEDSRPTTHDDPTFAVHDTVFYCVAN
MMAR_1928|M.marinum_M PNSLVARMKKGAVLVDISIDQGGCFEDSRPTTHTSPTFTVHDTVFYCVAN
MUL_2154|M.ulcerans_Agy99 PNSLVARMKKGAVLVDISIDQGGCFEDSRPTTHTSPTFTVHDTVFYCVAN
MAB_3100|M.abscessus_ATCC_1997 SNSLVSQMKPGSVLVDIAIDQGGCFADSRPTTHDEPTYTVHDSVFYCVAN
MSMEG_2659|M.smegmatis_MC2_155 TNSTVAHMKPGAVLVDIAIDQGGCFEDSRPTTHDEPTFKVHDTIFYCVAN
TH_1286|M.thermoresistible__bu SNSTVAGMRPGAVVVDVAIDQGGCFEDSRPTTFDDPTYHVHDTVFCCIGN
.** *: *: *:*:**::******* .*:***. **: ***::* *:.*
Mflv_4017|M.gilvum_PYR-GCK MPGAVPRTSTYALTNATLPYVLKLAGRGWRDACRADPALAKGLSTHEGAL
Mvan_2355|M.vanbaalenii_PYR-1 MPGAVPRTSTWALTNATMPYVLKIADKGWRDACRADAALANGLSTHEGAL
Rv2780|M.tuberculosis_H37Rv MPASVPKTSTYALTNATMPYVLELADHGWRAACRSNPALAKGLSTHEGAL
Mb2803|M.bovis_AF2122/97 MPASVPKTSTYALTNATMPYVLELADHGWRAACRSNPALAKGLSTHEGAL
MLBr_01532|M.leprae_Br4923 MPSAVPKTSTFALTNATMPYILKLADQGWQEACQSDPALAKGLSTHNGAL
MAV_3674|M.avium_104 MPSAVPKTSTVALTNATMPYVLALADRGWRAACRSDHALAKGLSTHDGTL
MMAR_1928|M.marinum_M MPGAAPRTSTYALTNATMPYVLKLADLGWQAACLSDSALAKGLSTHEGAL
MUL_2154|M.ulcerans_Agy99 MPGAAPRTSTYALTNATMPYVLKLADLGWQAACLSDSALAKGLSTHEGAL
MAB_3100|M.abscessus_ATCC_1997 MPGAVPRTSTFALTNATMPYVLALADKGWRQACRDDAALAQGLSTHEGAL
MSMEG_2659|M.smegmatis_MC2_155 MPGAVPRTSTFALTNSTMPYVLKLADKGWQAACASDSALAKGLSTHDGKL
TH_1286|M.thermoresistible__bu LPGAVPRTSTFALVNATLPYVVRLADHGWRAACVADPALARGLATHFGEL
:*.:.*:*** **.*:*:**:: :*. **: ** : ***.**:** * *
Mflv_4017|M.gilvum_PYR-GCK LSEQVAADLDVPYTDPAELLA--
Mvan_2355|M.vanbaalenii_PYR-1 LSDEVAADLDLPYTDPADVLA--
Rv2780|M.tuberculosis_H37Rv LSERVATDLGVPFTEPASVLA--
Mb2803|M.bovis_AF2122/97 LSERVATDLGVPFTEPASVLA--
MLBr_01532|M.leprae_Br4923 LSEQVATDLGLAFTEPASVLA--
MAV_3674|M.avium_104 LSQQVAADLGLPCTDPAGVLQQD
MMAR_1928|M.marinum_M LADHVAADLDLPFTDPANVLG--
MUL_2154|M.ulcerans_Agy99 LADHVAADLDLPFTDPASVLG--
MAB_3100|M.abscessus_ATCC_1997 LNTHVAHDLGLAFTDPAEVLG--
MSMEG_2659|M.smegmatis_MC2_155 LSEAVAKDLDLPFTDAAQFLA--
TH_1286|M.thermoresistible__bu FSASVAGDLGLPYSDPATLLG--
: ** **.:. ::.* .*