For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MAASPRWQCDRAAGASGAIQFHGVITDEDGRPVPNVLIETWHAAGDDDNAATKIGRLRCDGRNYTLPKSV ITDQQGRYSMRMVPPRAPEHGAAPFIAVSVHTRGLLDRLITRAYLPGCHLAKDPLLRRLPAERRQALIAT RDDIGLRFDITLRPAHGVRVETFVEPQQEAAT
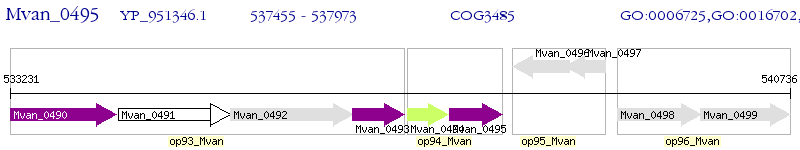
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_0495 | - | - | 100% (172) | intradiol ring-cleavage dioxygenase |
| M. vanbaalenii PYR-1 | Mvan_0561 | - | 4e-30 | 49.30% (142) | protocatechuate 3,4-dioxygenase, alpha subunit |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0655 | - | 1e-97 | 98.84% (172) | intradiol ring-cleavage dioxygenase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_3505 | pcaG | 4e-19 | 38.03% (142) | protocatechuate 3,4-dioxygenase, alpha subunit |
| M. smegmatis MC2 155 | MSMEG_2602 | pcaG | 1e-32 | 51.43% (140) | protocatechuate 3,4-dioxygenase, alpha subunit |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_3505|M.avium_104 -------------------MPYPGDARLVDDGDPRAVRLHGTVYDGVGAA
MSMEG_2602|M.smegmatis_MC2_155 MSTLSATPGQTVGPFYGYALPIPGGNELVPPGSPGAIRLHGLVTDGAGAP
Mvan_0495|M.vanbaalenii_PYR-1 ------------------MAASPRWQCDRAAGASGAIQFHGVITDEDGRP
Mflv_0655|M.gilvum_PYR-GCK ------------------MAASPRWQCDRAAGASGAIQFHGVITDEDGRP
. * * . *:::** : * * .
MAV_3505|M.avium_104 VPDALVELWQPDGGG-RIVRQAGSLRRDPAVFTGWGRCATGEDGGYGFTT
MSMEG_2602|M.smegmatis_MC2_155 VPDALLEIWQADADG-VVPTKTGSLRRDGWTFTGWGRAATDGAGRYSFTT
Mvan_0495|M.vanbaalenii_PYR-1 VPNVLIETWHAAGDDDNAATKIGRLRCDGRNYTLPKSVITDQQGRYSMRM
Mflv_0655|M.gilvum_PYR-GCK VPNVLIETWHAAGDDDNAATKIGRPRCDGRNYTRPKSVITDQQGRYSMRM
**:.*:* *:. ... : * * * :* *. * *.:
MAV_3505|M.avium_104 LAPGSVIDGRTPFFALTVFARGLLNRLFTRAYLPGAGPDTDPLLARVAPE
MSMEG_2602|M.smegmatis_MC2_155 VEPGVSQTGSAPFIAMTVFARGLLNRLFTRVYLPGEHVATDPLLSSLPSG
Mvan_0495|M.vanbaalenii_PYR-1 VPPRAPEHGAAPFIAVSVHTRGLLDRLITRAYLPGCHLAKDPLLRRLPAE
Mflv_0655|M.gilvum_PYR-GCK VPPRAPEHGAAPFIAVSVHTRGLLDRLITRAYLPGCHLAKDPLLRRLPAE
: * * :**:*::*.:****:**:**.**** .**** :..
MAV_3505|M.avium_104 RRTTLLCVAENGGRAYRFDIRLQ---GPGETVFLAYRTDGR-
MSMEG_2602|M.smegmatis_MC2_155 RRDTLIAVPDEHG--LTFDIRLQ---GDRETVFLRFPGHTP-
Mvan_0495|M.vanbaalenii_PYR-1 RRQALIATRDDIG--LRFDITLRPAHGVRVETFVEPQQEAAT
Mflv_0655|M.gilvum_PYR-GCK RRQALIATRDDIG--LRFDITLRPAHGVRVETFVEPQQEAAT
** :*:.. :: * *** *: * .*: .