For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
NTSLRRISVMIMALVVVLLANATLTQVFTADGLRSDPRNQRVLLDEYSRQRGQISAGGQLLAYSVSTQGR FRFLRVYPDPMIYAPVTGFYSLSYSSTGLERAEDTILNGSDERLFGRRLADFFTGRDPRGGTVATTINPQ VQRAAWDAMEKGCDGPCTGSVVALEPSTGKILALVSAPSYDPNLLATHDVAEQAEAWQRLRDDPGSPLLN RAISEAYPPGSTFKVITTAAALQAGATPDTQLTAAPQIALPDSTATLQNFGGVSCGAGPTTSLRNAFAHS CNTAFVQLGIDTGADPLRSTAGDFGVGAVPPAIPLQVAESTVGPIADDAALGMSSIGQRDVALTPLQNAM IAAAVANKGVMMRPYLVESLKGPDLANIATTAPTEERRAVTEQVADTLTDLMVAAEQVTQQKGAIAGVQI ASKTGTAEHGTDPRNTPPHAWYIAFAPAQAPKVAVAVLVENGGNRLSATGGALAAPIGRATIAAALREAS *
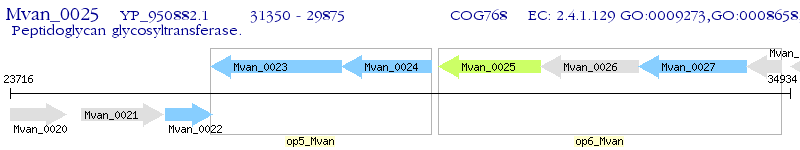
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_0025 | - | - | 100% (491) | penicillin-binding protein, transpeptidase |
| M. vanbaalenii PYR-1 | Mvan_4630 | - | 5e-21 | 29.77% (346) | penicillin-binding protein, transpeptidase |
| M. vanbaalenii PYR-1 | Mvan_3529 | - | 3e-19 | 26.15% (436) | peptidoglycan glycosyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0016c | pbpA | 0.0 | 78.69% (488) | penicillin-binding protein PbpA |
| M. gilvum PYR-GCK | Mflv_0810 | - | 0.0 | 87.98% (491) | penicillin-binding protein, transpeptidase |
| M. tuberculosis H37Rv | Rv0016c | pbpA | 0.0 | 78.69% (488) | penicillin-binding protein PbpA |
| M. leprae Br4923 | MLBr_00018 | pbpA | 0.0 | 77.71% (489) | putative penicillin-binding protein |
| M. abscessus ATCC 19977 | MAB_0035c | - | 0.0 | 69.44% (504) | penicillin-binding protein PbpA |
| M. marinum M | MMAR_0018 | pbpA | 0.0 | 80.53% (488) | penicillin-binding protein PbpA |
| M. avium 104 | MAV_0020 | - | 0.0 | 77.71% (489) | PbpA protein |
| M. smegmatis MC2 155 | MSMEG_0031 | - | 0.0 | 83.10% (491) | penicillin binding protein transpeptidase domain-containing |
| M. thermoresistible (build 8) | TH_3201 | - | 2e-19 | 26.95% (397) | PUTATIVE glycosyl transferase, family 51 |
| M. ulcerans Agy99 | MUL_0020 | pbpA | 0.0 | 80.12% (488) | penicillin-binding protein PbpA |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_0025|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_0810|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_0031|M.smegmatis_MC2_155 --------------------------------------------------
Mb0016c|M.bovis_AF2122/97 --------------------------------------------------
Rv0016c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_0018|M.marinum_M --------------------------------------------------
MUL_0020|M.ulcerans_Agy99 --------------------------------------------------
MAV_0020|M.avium_104 --------------------------------------------------
MLBr_00018|M.leprae_Br4923 --------------------------------------------------
MAB_0035c|M.abscessus_ATCC_199 --------------------------------------------------
TH_3201|M.thermoresistible__bu MRLSTLATTAAAVALTGTVGGLATGPAARSRWFARLKKPGFQPPAQAFPI
Mvan_0025|M.vanbaalenii_PYR-1 -----------------------------------------------MNT
Mflv_0810|M.gilvum_PYR-GCK -----------------------------------------------MNT
MSMEG_0031|M.smegmatis_MC2_155 -----------------------------------------------MNT
Mb0016c|M.bovis_AF2122/97 -----------------------------------------------MNA
Rv0016c|M.tuberculosis_H37Rv -----------------------------------------------MNA
MMAR_0018|M.marinum_M -----------------------------------------------MNA
MUL_0020|M.ulcerans_Agy99 -----------------------------------------------MNA
MAV_0020|M.avium_104 -----------------------------------------------MNA
MLBr_00018|M.leprae_Br4923 -----------------------------------------------MNT
MAB_0035c|M.abscessus_ATCC_199 -----------------------------------------------MNT
TH_3201|M.thermoresistible__bu VWPILYTDIAGVSAATIDRLXXXPGEPPEEPANWFDRINWRWVRRGLFAG
:
Mvan_0025|M.vanbaalenii_PYR-1 SLRRISVMIMALVVVLLAN---------------------ATLTQVFTAD
Mflv_0810|M.gilvum_PYR-GCK SLRRISVMIMALIVVLLAN---------------------ATLTQVFTAD
MSMEG_0031|M.smegmatis_MC2_155 SLRRVAVAIMVLIVLLLAN---------------------ATVTQVFAAD
Mb0016c|M.bovis_AF2122/97 SLRRISVTVMALIVLLLLN---------------------ATMTQVFTAD
Rv0016c|M.tuberculosis_H37Rv SLRRISVTVMALIVLLLLN---------------------ATMTQVFTAD
MMAR_0018|M.marinum_M SLRRISVTVMALIVLLLLN---------------------ATMTQVFTAD
MUL_0020|M.ulcerans_Agy99 SLRRISVTVMALIVLLLLN---------------------ATMTPVFTAD
MAV_0020|M.avium_104 SLRRISVTVMALIVLLLLN---------------------ATMTQVFAAD
MLBr_00018|M.leprae_Br4923 SLRRISVTVMALIVLLLLN---------------------ATVTQVFTAD
MAB_0035c|M.abscessus_ATCC_199 SIRRVSLAVMGLVVLLLAN---------------------ATITQVFTAP
TH_3201|M.thermoresistible__bu AVALLVLPIITFVMAYLIVDVPRPGDIRTNQVSTILASDGSELARIVPPE
:: : : :: ::: * : :: :...
Mvan_0025|M.vanbaalenii_PYR-1 GLRSDP----------------------RNQRVLLDEYSRQRGQISAGGQ
Mflv_0810|M.gilvum_PYR-GCK GLRSDP----------------------RNQRVLLDEYSRQRGQISAGGQ
MSMEG_0031|M.smegmatis_MC2_155 GLRADP----------------------RNQRVLLDEYSRQRGQITAGGQ
Mb0016c|M.bovis_AF2122/97 GLRADP----------------------RNQRVLLDEYSRQRGQITAGGQ
Rv0016c|M.tuberculosis_H37Rv GLRADP----------------------RNQRVLLDEYSRQRGQITAGGQ
MMAR_0018|M.marinum_M GLRADP----------------------RNQRVLLDEYSRQRGQITAGGQ
MUL_0020|M.ulcerans_Agy99 GLRADP----------------------RNQRVLLDEYSRQRGQITAGGQ
MAV_0020|M.avium_104 SLRADP----------------------RNQRVLLDEYSRQRGQIVAGGQ
MLBr_00018|M.leprae_Br4923 GLRADP----------------------RNQRVLLDEYSRQRGQITASGQ
MAB_0035c|M.abscessus_ATCC_199 GLRADP----------------------RNQRILLDEYSRQRGQISASGQ
TH_3201|M.thermoresistible__bu GNRIDVNINQIPVHVRDAVMAAEDRDFYSNPGFSFTGFARAIGNNIFGGD
. * * * . : ::* *: .*:
Mvan_0025|M.vanbaalenii_PYR-1 LLAYSVSTQGRFR--------------------------FLRVYPDPMIY
Mflv_0810|M.gilvum_PYR-GCK LLAYSVSTEGRFR--------------------------FLRVYPNPMAY
MSMEG_0031|M.smegmatis_MC2_155 LLAYSVSTDGRFR--------------------------YLRVYPNPQAY
Mb0016c|M.bovis_AF2122/97 LLAYSVATDGRFR--------------------------FLRVYPNPEVY
Rv0016c|M.tuberculosis_H37Rv LLAYSVATDGRFR--------------------------FLRVYPNPEVY
MMAR_0018|M.marinum_M LLAYSVATDSRFR--------------------------FLRVYPNPEVY
MUL_0020|M.ulcerans_Agy99 LLAYSVATDGRFR--------------------------FLRVYPNPEVY
MAV_0020|M.avium_104 LLAYSVATDNRFR--------------------------FLRVYPNPAQY
MLBr_00018|M.leprae_Br4923 LLAYSVATDNRFR--------------------------FLRVYPNPAVY
MAB_0035c|M.abscessus_ATCC_199 LLASSVATDSRFR--------------------------FLRQYADPEVY
TH_3201|M.thermoresistible__bu VQGGSTITQQYVKNALVGSERVGVRGLIRKAKELVISTKMSRQWSKDQVM
: . *. *: .: * :..
Mvan_0025|M.vanbaalenii_PYR-1 APVTGFYSLSYSSTGLERAEDTILNGSDERLFGRRLADFFTGRDPRGGTV
Mflv_0810|M.gilvum_PYR-GCK APVTGFYSLTYSSSGLERAEDTILNGSDERLFGRRLADFFTGRDPRGGNV
MSMEG_0031|M.smegmatis_MC2_155 APVTGFYSLGYSSTGLERAEDAVLNGSDERLFGRRLADFFTGRDPRGGNV
Mb0016c|M.bovis_AF2122/97 APVTGFYSLRYSSTALERAEDPILNGSDRRLFGRRLADFFTGRDPRGGNV
Rv0016c|M.tuberculosis_H37Rv APVTGFYSLRYSSTALERAEDPILNGSDRRLFGRRLADFFTGRDPRGGNV
MMAR_0018|M.marinum_M APVTGFYSLRYSSTGLERAEDPLLNGSDQRLFGRRLADFFTGRDPRGGNV
MUL_0020|M.ulcerans_Agy99 APVTGFYSLRYSSTGLERAEDPLLNGSDQRLFGRRLADFFTGRDPRGGNV
MAV_0020|M.avium_104 APVTGFYSLRYSSTGLERAEDPLLNGSDERLFGRRLADFFTGRDPRGANV
MLBr_00018|M.leprae_Br4923 APITGFYSLRYSSTGLEQAEDALLNGSDERLFGRRLADFFTGRDPRGGNV
MAB_0035c|M.abscessus_ATCC_199 APVTGFYSLNYSSTGLERAEDTMLNGSDPRLFGRRLADFFTGRDPRGGSV
TH_3201|M.thermoresistible__bu QSYLNIIYFGRGAYGVAAASRAYFDKPVEELN-VAEGALLAALIQRPSTL
. .: : .: .: *. . :: . .* . :::. * ..:
Mvan_0025|M.vanbaalenii_PYR-1 ATTINPQVQRAAWDAMEKGCDG-----PCTGSVVALEPSTGKILALVSAP
Mflv_0810|M.gilvum_PYR-GCK STTINPDVQQAAWDAMEDGCNG-----PCKGSVVALEPSTGKILAMVSAP
MSMEG_0031|M.smegmatis_MC2_155 DTTIKPQVQQAAWDAMQNGCDG-----PCRGSVVALEPSTGKILAMVSAP
Mb0016c|M.bovis_AF2122/97 DTTINPRIQQAGWDAMQQGCYG-----PCKGAVVALEPSTGKILALVSSP
Rv0016c|M.tuberculosis_H37Rv DTTINPRIQQAGWDAMQQGCYG-----PCKGAVVALEPSTGKILALVSSP
MMAR_0018|M.marinum_M DTTINPRIQQAGWDAMQQGCGG-----PCKGAVVALEPSTGKILALVSSP
MUL_0020|M.ulcerans_Agy99 DTAINPRIQQAGWDAMQQGCGG-----PCKGAVVALEPSTGKILALVSSP
MAV_0020|M.avium_104 DTTIRPRVQQAAWDGMQQGCGGP----PCKGAVVALEPSTGKILAMVSSP
MLBr_00018|M.leprae_Br4923 DTTINPRVQQTGWDAMQQGCGGS----PCKGAVVALEPSTGKILAMVSTP
MAB_0035c|M.abscessus_ATCC_199 VTTIRPDVQQAAYDAMHRGCKD-----GCRGAVVALEPSTGKILAMVSTP
TH_3201|M.thermoresistible__bu DPAADPQGAETRWNWVLDGMVEIGALSPQERAAQVFPPTVPPELARSQNQ
.: * .: :: : * :. .: *:. ** .
Mvan_0025|M.vanbaalenii_PYR-1 SYDPN----------------------------LLATHDVAEQAEAWQRL
Mflv_0810|M.gilvum_PYR-GCK SYDPN----------------------------LLASHNIDEQAQTWQAL
MSMEG_0031|M.smegmatis_MC2_155 SYDPN----------------------------LLATHDLAAQADAWEKL
Mb0016c|M.bovis_AF2122/97 SYDPN----------------------------LLASHNPEVQAQAWQRL
Rv0016c|M.tuberculosis_H37Rv SYDPN----------------------------LLASHNPEVQAQAWQRL
MMAR_0018|M.marinum_M SYDPN----------------------------LLASHDPEAQAEAWKDL
MUL_0020|M.ulcerans_Agy99 SYDPN----------------------------LLASHDPEAQAEAWKDL
MAV_0020|M.avium_104 SYDPN----------------------------LLSSHDPEVQAQAWQRL
MLBr_00018|M.leprae_Br4923 SYDPN----------------------------LLASHNPEEQAQAWRRL
MAB_0035c|M.abscessus_ATCC_199 SYDPN----------------------------LLASQDPEKENAAWKRL
TH_3201|M.thermoresistible__bu TTGPNGLIERQVTRELLELFNISEQALNTEGLQITTTIDPQAQQAAVDAV
: .** : :: : : : :
Mvan_0025|M.vanbaalenii_PYR-1 R-----DDPGSPLLNRAIS----------------------EAYPPGSTF
Mflv_0810|M.gilvum_PYR-GCK R-----DNPDSPLLNRAIS----------------------ETYPPGSTF
MSMEG_0031|M.smegmatis_MC2_155 R-----DDPQSPLLNRAIS----------------------ETYPPGSTF
Mb0016c|M.bovis_AF2122/97 G-----DNPASPLTNRAIS----------------------ETYPPGSTF
Rv0016c|M.tuberculosis_H37Rv G-----DNPASPLTNRAIS----------------------ETYPPGSTF
MMAR_0018|M.marinum_M R-----DDPSSPLTNRAIS----------------------ETYPPGSTF
MUL_0020|M.ulcerans_Agy99 R-----DDPSSALTNRAIS----------------------ETYPPGSTF
MAV_0020|M.avium_104 R-----DDPDNPMTNRAIS----------------------ETYPPGSTF
MLBr_00018|M.leprae_Br4923 H-----DDPNSPLINRAIS----------------------ETYPPGSTF
MAB_0035c|M.abscessus_ATCC_199 T-----EDPAQPMLNRAIA----------------------QTYPPGSTF
TH_3201|M.thermoresistible__bu ETYLEGQDPDMRAAVVSIDPRTGAVKAYYGGSDANGYDFAQAAVPTGSSF
::* :* : *.**:*
Mvan_0025|M.vanbaalenii_PYR-1 KVITTAAALQAGATPDTQLTAAPQIALPDSTATLQNFGGVSCGAGPTTSL
Mflv_0810|M.gilvum_PYR-GCK KVLTTAAALEAGATPQTQLTAAPQIPLPNSTATLENFGGTACGGGPTATL
MSMEG_0031|M.smegmatis_MC2_155 KVITTAAALQAGARPQTQLTSAPRTPLPDSTATLENFGGAPCGPGPTVSL
Mb0016c|M.bovis_AF2122/97 KVITTAAALAAGATETEQLTAAPTIPLPGSTAQLENYGGAPCGDEPTVSL
Rv0016c|M.tuberculosis_H37Rv KVITTAAALAAGATETEQLTAAPTIPLPGSTAQLENYGGAPCGDEPTVSL
MMAR_0018|M.marinum_M KVITTAAALQAGATENDQLTAAANIPLPDSTATLENYGGAPCDSGQTVSL
MUL_0020|M.ulcerans_Agy99 KVITTAAALQAGATENDQLTAAANIPLPDSTATLENYGGAPCGSGQTVSL
MAV_0020|M.avium_104 KVITTAAALQAGASDTEQLTAAPSIPLPNSTATLENYGGQACGNDPTVSL
MLBr_00018|M.leprae_Br4923 KVITTTAALQAGATTSDQLTAEPTIPLPGSTATLENYGGVSCGPGPTVSL
MAB_0035c|M.abscessus_ATCC_199 KVITTAAALADGKTVNTEVTASPTIFLPGSNATLENYNRASCGGGDTTSL
TH_3201|M.thermoresistible__bu KVFALVAALEQGMGLGYQVDSSP---LTVNGIEITNVEGSSCG---RCNI
**:: .*** * :: : . *. . : * .*. .:
Mvan_0025|M.vanbaalenii_PYR-1 RNAFAHSCNTAFVQLGIDTG--ADPLRSTAGDFGVGAVPPAIPLQVAEST
Mflv_0810|M.gilvum_PYR-GCK EYAFAHSCNTAFVQLGIDTG--ADALRSTAVGFGVDTAPPAIPLQVAEST
MSMEG_0031|M.smegmatis_MC2_155 QEAFAKSCNTAFVELGLSTG--TDKLKAMAQAFGLDTPPPAIPLQVAEST
Mb0016c|M.bovis_AF2122/97 REAFVKSCNTAFVQLGIRTG--ADALRSMARAFGLDSPPRPTPLQVAEST
Rv0016c|M.tuberculosis_H37Rv REAFVKSCNTAFVQLGIRTG--ADALRSMARAFGLDSPPRPTPLQVAEST
MMAR_0018|M.marinum_M REAFAKSCNTAFVQLGISTG--AAALRSMAHAFGLDTPPSAIPLQVAEST
MUL_0020|M.ulcerans_Agy99 REAFAKSCNTAFVQLGISTG--AAALRSMAHAFGLDTPPSAIPLQVAEST
MAV_0020|M.avium_104 QQAFALSCNTAFVQLGILTG--ADALRSMARSFGLDSTPSVIPLQVAEST
MLBr_00018|M.leprae_Br4923 SQAFAMSCNTAFVQLGLLIG--ADALRSMAHSFGLDSNPNVIPLQVAEST
MAB_0035c|M.abscessus_ATCC_199 KEAFARSCNTAFAKLGAEQLK-RDKLVKAAQAFGLDRELDSIPLQVATST
TH_3201|M.thermoresistible__bu AEALKRSLNTSFYRLMLELKNGPEDVADAAHRAGIAESIPGIEHTLSEDG
*: * **:* .* : * *: :: .
Mvan_0025|M.vanbaalenii_PYR-1 VGPIADDAALGMSSIGQRDVALTPLQNAMIAAAVANKGVMMRPYLVESLK
Mflv_0810|M.gilvum_PYR-GCK IGPIVDDAALGMTSIGQRDVAMTPLQNAMVAATVANNGVTMTPYLVESLK
MSMEG_0031|M.smegmatis_MC2_155 TGPIVDAAALGMSSIGQRDVALTPLQNAQVAATIANDGIAMRPYLVESLK
Mb0016c|M.bovis_AF2122/97 VGPIPDSAALGMTSIGQKDVALTPLANAEIAATIANGGITMRPYLVGSLK
Rv0016c|M.tuberculosis_H37Rv VGPIPDSAALGMTSIGQKDVALTPLANAEIAATIANGGITMRPYLVGSLK
MMAR_0018|M.marinum_M VGPIPDAAALGMSSIGQKDVALTPLQNAQIAATIANGGVVMRPYLVDSLK
MUL_0020|M.ulcerans_Agy99 VGPIPDAAALGMSSIGQKDVALTPLQNAQIAATIANGGVVMRPYLVDGLK
MAV_0020|M.avium_104 IGIIPDAAALGMSSIGQKDVALTPLQNAEIAATIANGGVTMQPYLVDSLK
MLBr_00018|M.leprae_Br4923 VGVIPDAAALGMSSIGQRDVALTPLQNAQIAATIANAGVTMQPYLIDNLK
MAB_0035c|M.abscessus_ATCC_199 VGPMSDDAAVAMSSIGQKDVAMTPLQNAMVAAAVANKGVLMQPHLVDSLK
TH_3201|M.thermoresistible__bu KGGPPNNGIVLGQYQSR------PIDMASAYATLANSGVYHRPHFVQKVV
* : . : .: *: * *::** *: *::: :
Mvan_0025|M.vanbaalenii_PYR-1 GPDLANIATTAPTEE--RRAVTEQVADTLTDLMVAAEQVTQQKGAIAGVQ
Mflv_0810|M.gilvum_PYR-GCK GPDLANIATTSPTEE--RRAVPEKVADTLTDLMVAAEQVTQQKGAIAGVQ
MSMEG_0031|M.smegmatis_MC2_155 GPDLATISTTTPEQE--RRAVSPQVAATLTDLMVAAEQVTQQKGAIAGVQ
Mb0016c|M.bovis_AF2122/97 GPDLANISTTVGYQQ--RRAVSPQVAAKLTELMVGAEKVAQQKGAIPGVQ
Rv0016c|M.tuberculosis_H37Rv GPDLANISTTVGYQQ--RRAVSPQVAAKLTELMVGAEKVAQQKGAIPGVQ
MMAR_0018|M.marinum_M GPDLANISTTTPHQQ--RRAVSPQVAAKLTELMVGAEQVAQQKGAIPGVQ
MUL_0020|M.ulcerans_Agy99 GPDLANISTTAPHQQ--RRAVSPQVAAKLTELMVGAEQVAQQKGAIPGVQ
MAV_0020|M.avium_104 GPDLTTISTTTPYEQ--RRAVSPQVAAKLTELMVGAEKVAQQKGAIPGVQ
MLBr_00018|M.leprae_Br4923 GPDLANIRTTTSYQQ--HRAVSPQIAATLTELMVGAEKVTQQKGAIPGVQ
MAB_0035c|M.abscessus_ATCC_199 GPDLSNLSTTTPDDI--GQAVTPAIANTLTELMIASEQRTAQTGALPGVQ
TH_3201|M.thermoresistible__bu NSEGQVLFDADQRDDDGEQRIEKDVADNVTAAMQPIAAYSNGHALAGGRP
..: : : : : : :* .:* * : . *
Mvan_0025|M.vanbaalenii_PYR-1 IASKTGTAEHGTDPRNTPPHAWYIAFAPAQ--------------------
Mflv_0810|M.gilvum_PYR-GCK IASKTGTAEHGTDPRNTPPHAWYIAFAPAQ--------------------
MSMEG_0031|M.smegmatis_MC2_155 IASKTGTAEHGTDPRNTPPHAWYIAFAPAQ--------------------
Mb0016c|M.bovis_AF2122/97 IASKTGTAEHGTDPRHTPPHAWYIAFAPAQ--------------------
Rv0016c|M.tuberculosis_H37Rv IASKTGTAEHGTDPRHTPPHAWYIAFAPAQ--------------------
MMAR_0018|M.marinum_M IASKTGTAEHGTDPRHTPPHAWYIAFAPAQ--------------------
MUL_0020|M.ulcerans_Agy99 IASKTGTAEHGTDPRHTPPHAWYIAFAPAQ--------------------
MAV_0020|M.avium_104 IASKTGTAEHGSDPRHTPPHAWYIAFAPAQ--------------------
MLBr_00018|M.leprae_Br4923 IASKTGTAEHGTDPRHTPPHAWYIAFAPVQ--------------------
MAB_0035c|M.abscessus_ATCC_199 IASKTGTAEHGTDPRNTPPHAWYIAFAPAQGTTTDP--------------
TH_3201|M.thermoresistible__bu SAAKTGTHQLGDTGENR--DAWMVGYTPSLSTAVWVGTVDGNKPLRTKWG
*:**** : * .: .** :.::*
Mvan_0025|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_0810|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_0031|M.smegmatis_MC2_155 --------------------------------------------------
Mb0016c|M.bovis_AF2122/97 --------------------------------------------------
Rv0016c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_0018|M.marinum_M --------------------------------------------------
MUL_0020|M.ulcerans_Agy99 --------------------------------------------------
MAV_0020|M.avium_104 --------------------------------------------------
MLBr_00018|M.leprae_Br4923 --------------------------------------------------
MAB_0035c|M.abscessus_ATCC_199 --------------------------------------------------
TH_3201|M.thermoresistible__bu GKIYGSGLPSDIWKATMDGALKGTERESFPKPGEIGGYAGVPQAPSPQRT
Mvan_0025|M.vanbaalenii_PYR-1 -------------------APKVAVAVLVENG------------------
Mflv_0810|M.gilvum_PYR-GCK -------------------APTVAVAVLVENG------------------
MSMEG_0031|M.smegmatis_MC2_155 -------------------DPKVAVAVLVEDG------------------
Mb0016c|M.bovis_AF2122/97 -------------------APKVAVAVLVENG------------------
Rv0016c|M.tuberculosis_H37Rv -------------------APKVAVAVLVENG------------------
MMAR_0018|M.marinum_M -------------------APKVAVAVLVENG------------------
MUL_0020|M.ulcerans_Agy99 -------------------APKVAVAVLVENG------------------
MAV_0020|M.avium_104 -------------------TPKVAVAVLVENG------------------
MLBr_00018|M.leprae_Br4923 -------------------APKVAVAVLVEKG------------------
MAB_0035c|M.abscessus_ATCC_199 --------------QATAVTPKVAIAVLVEDG------------------
TH_3201|M.thermoresistible__bu SDDDGTTTVTPPSETVIQPSVEVAPGITIPLGXAVSAVVRVRDAVVRPHL
** .: : *
Mvan_0025|M.vanbaalenii_PYR-1 -----------------------------------------GNR-LSATG
Mflv_0810|M.gilvum_PYR-GCK -----------------------------------------GDR-LGATG
MSMEG_0031|M.smegmatis_MC2_155 -----------------------------------------GDR-LSATG
Mb0016c|M.bovis_AF2122/97 -----------------------------------------ADR-LSATG
Rv0016c|M.tuberculosis_H37Rv -----------------------------------------ADR-LSATG
MMAR_0018|M.marinum_M -----------------------------------------ADR-LSATG
MUL_0020|M.ulcerans_Agy99 -----------------------------------------ADR-LSATG
MAV_0020|M.avium_104 -----------------------------------------ADR-LSATG
MLBr_00018|M.leprae_Br4923 -----------------------------------------ADS-LFATG
MAB_0035c|M.abscessus_ATCC_199 -----------------------------------------GDRALAATG
TH_3201|M.thermoresistible__bu VAEPPLTGRPDGTSRPGRRAPVRRCRNVDFRRDIGAPCAFHRDRRWLPAD
: .:.
Mvan_0025|M.vanbaalenii_PYR-1 GALAAPIGRATIAAALREAS------------------------------
Mflv_0810|M.gilvum_PYR-GCK GALAAPIGRATIAAALRGAS------------------------------
MSMEG_0031|M.smegmatis_MC2_155 GALAAPIGRATIAAALREGS------------------------------
Mb0016c|M.bovis_AF2122/97 GALAAPIGRAVIEAALQGEP------------------------------
Rv0016c|M.tuberculosis_H37Rv GALAAPIGRAVIEAALQGEP------------------------------
MMAR_0018|M.marinum_M GALAAPIGRAVIEAALQGEP------------------------------
MUL_0020|M.ulcerans_Agy99 GALAAPIGQAVIEAALQGEP------------------------------
MAV_0020|M.avium_104 GALAAPIGRAVIEAALQGGP------------------------------
MLBr_00018|M.leprae_Br4923 GALAAPIGRAVIEAALQGGA------------------------------
MAB_0035c|M.abscessus_ATCC_199 GSVAAPIGRQVIAAALQGAS------------------------------
TH_3201|M.thermoresistible__bu AVRAEPLGRGSPQRTGPGRAGGLLSGAAVRGTGVPSEPAHGGSVPRRAGG
. * *:*: : .
Mvan_0025|M.vanbaalenii_PYR-1 ---------------------------------------------
Mflv_0810|M.gilvum_PYR-GCK ---------------------------------------------
MSMEG_0031|M.smegmatis_MC2_155 ---------------------------------------------
Mb0016c|M.bovis_AF2122/97 ---------------------------------------------
Rv0016c|M.tuberculosis_H37Rv ---------------------------------------------
MMAR_0018|M.marinum_M ---------------------------------------------
MUL_0020|M.ulcerans_Agy99 ---------------------------------------------
MAV_0020|M.avium_104 ---------------------------------------------
MLBr_00018|M.leprae_Br4923 ---------------------------------------------
MAB_0035c|M.abscessus_ATCC_199 ---------------------------------------------
TH_3201|M.thermoresistible__bu ADDGADAPGAGRAPGAQRRVRHHPGGPDGGPRQPAAVSPLRGTTR